5IHY
 
 | |
5ITG
 
 | |
5KNR
 
 | | E. coli HPRT in complexed with 9-[(N-phosphonoethyl-N-phosphonoethoxyethyl)-2-aminoethyl]-guanine | | Descriptor: | (2-{[2-(2-amino-6-oxo-3,6-dihydro-9H-purin-9-yl)ethyl][2-(2-phosphonoethoxy)ethyl]amino}ethyl)phosphonic acid, Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Eng, W.S, Keough, D.T, Hockova, D, Janeba, Z. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.864 Å) | | Cite: | Crystal Structures of Acyclic Nucleoside Phosphonates in Complex with Escherichia coli Hypoxanthine Phosphoribosyltransferase
Chemistryselect, 1, 2016
|
|
5KNU
 
 | | Crystal structure of E. coli hypoxanthine phosphoribosyltransferase in complexed with 9-[N,N-(Bis-3-phosphonopropyl)aminomethyl]-9-deazahypoxanthine | | Descriptor: | 3-[(4-oxidanylidene-3,5-dihydropyrrolo[3,2-d]pyrimidin-7-yl)methyl-(3-phosphonopropyl)amino]propylphosphonic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hypoxanthine-guanine phosphoribosyltransferase, ... | | Authors: | Eng, W.S, Keough, D.T, Baszczynski, O, Hockova, D, Janeba, Z. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Crystal Structures of Acyclic Nucleoside Phosphonates in Complex with Escherichia coli Hypoxanthine Phosphoribosyltransferase
Chemistryselect, 1, 2016
|
|
5KNX
 
 | | Crystal structure of E. coli hypoxanthine phosphoribosyltransferase in complexed with {[(2-[(Hypoxanthin-9H-yl)methyl]propane-1,3-diyl)bis(oxy)]bis- (methylene)}diphosphonic Acid | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION, [2-[(6-oxidanylidene-1~{H}-purin-9-yl)methyl]-3-(phosphonomethoxy)propoxy]methylphosphonic acid | | Authors: | Eng, W.S, Keough, D.T, Hockova, D, Janeba, Z, Guddat, L.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Acyclic Nucleoside Phosphonates in Complex with Escherichia coli Hypoxanthine Phosphoribosyltransferase
Chemistryselect, 1, 2016
|
|
5KNQ
 
 | | Crystal structure of Mycobacterium tuberculosis hypoxanthine guanine phosphoribosyltransferase in complex with [3S,4R]-(4-(Guanin-9-yl)pyrrolidin-3-yl)oxymethanephosphonic acid and pyrophosphate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Eng, W.S, Rejman, D, Keough, D.T, Guddat, L.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-09-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis hypoxanthine guanine phosphoribosyltransferase in complex with pyrrolidine nucleoside phosphonate
To Be Published
|
|
5KNY
 
 | | Crystal structure of Mycobacterium tuberculosis hypoxanthine guanine phosphoribosyltransferase in complex with (3-((3R,4R)-3-(Guanin-9-yl)-4-((S)-2-hydroxy-2-phosphonoethoxy)pyrrolidin-1-yl)-3-oxopropyl)phosphonic acid | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION, [3-[(3~{R},4~{R})-3-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-4-[(2~{S})-2-oxidanyl-2-phosphono-ethoxy]pyrrolidin-1-y l]-3-oxidanylidene-propyl]phosphonic acid | | Authors: | Eng, W.S, Rejman, D, Keough, D.T, Guddat, L.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis hypoxanthine guanine phosphoribosyltransferase in complex with pyrrolidine nucleoside phosphonate
To Be Published
|
|
5KNT
 
 | | Crystal structure of E. coli hypoxanthine phosphoribosyltransferase in complexed with 2-((2,3-Dihydroxypropyl)(2-(hypoxanthin-9-yl)ethyl)amino)ethylphosphonic acid | | Descriptor: | 2-[[(2~{S})-2,3-bis(oxidanyl)propyl]-[2-(6-oxidanylidene-1~{H}-purin-9-yl)ethyl]amino]ethylphosphonic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hypoxanthine-guanine phosphoribosyltransferase, ... | | Authors: | Eng, W.S, Keough, D.T, Hockova, D, Janeba, Z. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structures of Acyclic Nucleoside Phosphonates in Complex with Escherichia coli Hypoxanthine Phosphoribosyltransferase
Chemistryselect, 1, 2016
|
|
5KNV
 
 | | E coli hypoxanthine guanine phosphoribosyltransferase in complexed with 9-[(N-Phosphonoethyl-N-phosphonobutyl)-2-aminoethyl]-hypoxanthine | | Descriptor: | 2-[2-(6-oxidanylidene-1~{H}-purin-9-yl)ethyl-(4-phosphonobutyl)amino]ethylphosphonic acid, Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Eng, W.S, Keough, D.T, Hockova, D, Janeba, Z. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.861 Å) | | Cite: | Crystal Structures of Acyclic Nucleoside Phosphonates in Complex with Escherichia coli Hypoxanthine Phosphoribosyltransferase
Chemistryselect, 1, 2016
|
|
1WUE
 
 | | Crystal structure of protein GI:29375081, unknown member of enolase superfamily from enterococcus faecalis V583 | | Descriptor: | mandelate racemase/muconate lactonizing enzyme family protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-05 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1WUF
 
 | | Crystal structure of protein GI:16801725, member of Enolase superfamily from Listeria innocua Clip11262 | | Descriptor: | MAGNESIUM ION, hypothetical protein lin2664 | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-07 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1XBY
 
 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase E112D/T169A mutant with bound D-ribulose 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, MAGNESIUM ION, RIBULOSE-5-PHOSPHATE | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1XBZ
 
 | | Crystal structure of 3-keto-L-gulonate 6-phosphate decarboxylase E112D/R139V/T169A mutant with bound L-xylulose 5-phosphate | | Descriptor: | 3-Keto-L-Gulonate 6-Phosphate Decarboxylase, L-XYLULOSE 5-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1XFR
 
 | |
1X7B
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH ERB-041 | | Descriptor: | 2-(3-FLUORO-4-HYDROXYPHENYL)-7-VINYL-1,3-BENZOXAZOL-5-OL, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
1W9R
 
 | | Solution Structure of Choline Binding Protein A, Domain R2, the Major Adhesin of Streptococcus pneumoniae | | Descriptor: | CHOLINE BINDING PROTEIN A | | Authors: | Luo, R, Mann, B, Lewis, W.S, Rowe, A, Heath, R, Stewart, M.L, Hamburger, A.E, Bjorkman, P.J, Sivakolundu, S, Lacy, E.R, Tuomanen, E, Kriwacki, R.W. | | Deposit date: | 2004-10-15 | | Release date: | 2005-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Choline Binding Protein A, the Major Adhesin of Streptococcus Pneumoniae
Embo J., 24, 2005
|
|
1VQI
 
 | | GENE V PROTEIN MUTANT WITH ILE 47 REPLACED BY VAL 47 (I47V) | | Descriptor: | GENE V PROTEIN | | Authors: | Zhang, H, Skinner, M.M, Sandberg, W.S, Wang, A.H.-J, Terwilliger, T.C. | | Deposit date: | 1996-08-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Context dependence of mutational effects in a protein: the crystal structures of the V35I, I47V and V35I/I47V gene V protein core mutants.
J.Mol.Biol., 259, 1996
|
|
1VQF
 
 | | GENE V PROTEIN MUTANT WITH VAL 35 REPLACED BY ILE 35 AND ILE 47 REPLACED BY VAL 47 (V35I, I47V) | | Descriptor: | GENE V PROTEIN | | Authors: | Zhang, H, Skinner, M.M, Sandberg, W.S, Wang, A.H.-J, Terwilliger, T.C. | | Deposit date: | 1996-08-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Context dependence of mutational effects in a protein: the crystal structures of the V35I, I47V and V35I/I47V gene V protein core mutants.
J.Mol.Biol., 259, 1996
|
|
1X76
 
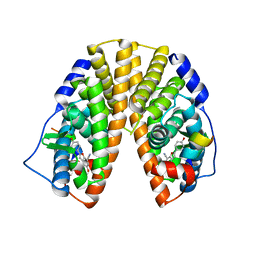 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH WAY-697 | | Descriptor: | 5-HYDROXY-2-(4-HYDROXYPHENYL)-1-BENZOFURAN-7-CARBONITRILE, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
1X7E
 
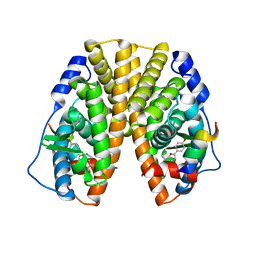 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR ALPHA COMPLEXED WITH WAY-244 | | Descriptor: | Estrogen receptor 1 (alpha), [5-HYDROXY-2-(4-HYDROXYPHENYL)-1-BENZOFURAN-7-YL]ACETONITRILE, steroid receptor coactivator-3 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
1YEY
 
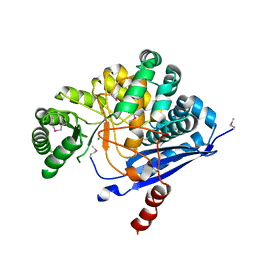 | | Crystal Structure of L-fuconate Dehydratase from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | L-fuconate dehydratase, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-28 | | Release date: | 2005-01-11 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of L-fuconate Dehydratase from Xanthomonas campestris pv. campestris str. ATCC 33913
To be Published
|
|
1XBX
 
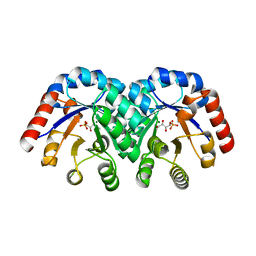 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase E112D/R139V/T169A mutant with bound D-ribulose 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, 5-O-phosphono-L-ribulose, MAGNESIUM ION, ... | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1X7J
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH GENISTEIN | | Descriptor: | Estrogen receptor beta, GENISTEIN, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Manas, E.S, Xu, Z.B, Unwalla, R.J, Somers, W.S. | | Deposit date: | 2004-08-14 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Understanding the Selectivity of Genistein for Human Estrogen Receptor-Beta Using X-Ray Crystallography and Computational Methods
Structure, 12, 2004
|
|
1XBV
 
 | | Crystal structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound D-ribulose 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, MAGNESIUM ION, RIBULOSE-5-PHOSPHATE | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1X7R
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR ALPHA COMPLEXED WITH GENISTEIN | | Descriptor: | Estrogen receptor 1 (alpha), GENISTEIN, steroid receptor coactivator-3 | | Authors: | Manas, E.S, Xu, Z.B, Unwalla, R.J, Somers, W.S. | | Deposit date: | 2004-08-16 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding the Selectivity of Genistein for Human Estrogen Receptor-Beta Using X-Ray Crystallography and Computational Methods
Structure, 12, 2004
|
|
