4WZ3
 
 | | Crystal structure of the complex between LubX/LegU2/Lpp2887 U-box 1 and Homo sapiens UBE2D2 | | Descriptor: | E3 ubiquitin-protein ligase LubX, Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Stogios, P.J, Quaile, A.T, Skarina, T, Nocek, B, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-18 | | Release date: | 2015-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
4WZ0
 
 | | Crystal structure of U-box 1 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris | | Descriptor: | E3 ubiquitin-protein ligase LubX | | Authors: | Stogios, P.J, Quaile, A.T, Skarina, T, Stein, A, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-18 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
4DCI
 
 | | Crystal structure of unknown funciton protein from Synechococcus sp. WH 8102 | | Descriptor: | SULFATE ION, uncharacterized protein | | Authors: | Chang, C, Marshall, N, Bearden, J, Palenik, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-17 | | Release date: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal structure of unknown function protein from Synechococcus sp. WH 8102
To be Published
|
|
4RGI
 
 | | Crystal Structure of KTSC Domain Protein YPO2434 from Yersinia pestis | | Descriptor: | GLYCEROL, SULFATE ION, Uncharacterized protein | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-31 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | Crystal Structure of KTSC Domain Protein YPO2434 from Yersinia pestis
To be Published
|
|
4RGK
 
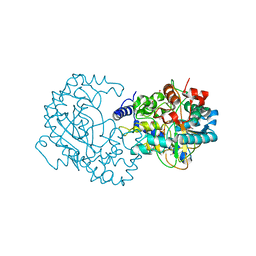 | | Crystal Structure of Putative Phytanoyl-CoA Dioxygenase Family Protein YbiU from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Putative Phytanoyl-CoA Dioxygenase Family Protein YbiU from Yersinia pestis
To be Published
|
|
4RI0
 
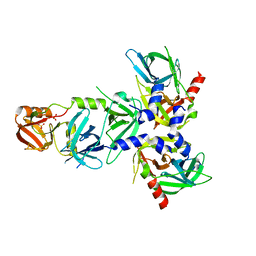 | | Serine Protease HtrA3, mutationally inactivated | | Descriptor: | PHOSPHATE ION, Serine protease HTRA3 | | Authors: | Osipiuk, J, Glaza, P, Wenta, T, Lipinska, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-03 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.272 Å) | | Cite: | Structural and Functional Analysis of Human HtrA3 Protease and Its Subdomains.
Plos One, 10, 2015
|
|
4XLT
 
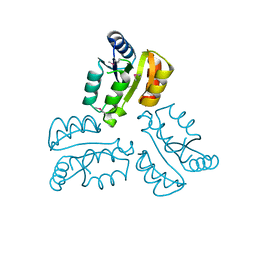 | | Crystal structure of response regulator receiver protein from Dyadobacter fermentans DSM 18053 | | Descriptor: | Response regulator receiver protein | | Authors: | Chang, C, Cuff, M, Holowicki, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-13 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of response regulator receiver protein from Dyadobacter fermentans DSM 18053
To Be Published
|
|
4XTK
 
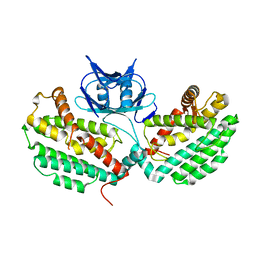 | | Structure of TM1797, a CAS1 protein from Thermotoga maritima | | Descriptor: | CRISPR-associated endonuclease Cas1 | | Authors: | Petit, P, Beloglazova, N, Skarina, T, Chang, C, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and nuclease activity of tm1797, a cas1 protein from thermotoga maritima
To Be Published
|
|
4RNL
 
 | | The crystal structure of a possible galactose mutarotase from Streptomyces platensis subsp. rosaceus | | Descriptor: | GLYCEROL, PHOSPHATE ION, possible galactose mutarotase | | Authors: | Tan, K, Li, H, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a possible galactose mutarotase from Streptomyces platensis subsp. rosaceus
To be Published
|
|
4TKT
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsF KS6 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | Authors: | Chang, C, Li, H, Endres, M, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4289 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4DGH
 
 | | Structure of SulP Transporter STAS Domain from Vibrio Cholerae Refined to 1.9 Angstrom Resolution | | Descriptor: | GLYCEROL, IODIDE ION, POTASSIUM ION, ... | | Authors: | Keller, J.P, Chang, C, Marshall, N, Bearden, J, Dallos, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-25 | | Release date: | 2012-02-08 | | Last modified: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of SulP Transporter STAS Domain from Vibrio Cholerae Refined to 1.9 Angstrom Resolution
To be Published
|
|
4DBX
 
 | | Crystal structure of aminoglycoside phosphotransferase APH(2")-ID/APH(2")-IVA | | Descriptor: | APH(2")-ID | | Authors: | Stogios, P.J, Minasov, G, Tan, K, Nocek, B, Singer, A.U, Evdokimova, E, Egorova, E, Di Leo, R, Li, H, Shakya, T, Wright, G.D, Savchenko, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-16 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | A small molecule discrimination map of the antibiotic resistance kinome.
Chem.Biol., 18, 2011
|
|
4RBS
 
 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 in the Complex with Hydrolyzed Meropenem | | Descriptor: | (2S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3-methyl-2H-pyrro le-5-carboxylic acid, ACETIC ACID, Beta-lactamase NDM-1, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 in the Complex with Hydrolyzed Meropenem
To be Published
|
|
4RD7
 
 | | The crystal structure of a Cupin 2 conserved barrel domain protein from Salinispora arenicola CNS-205 | | Descriptor: | Cupin 2 conserved barrel domain protein, GLYCEROL, SULFATE ION | | Authors: | Tan, K, Gu, M, Clancy, S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-09-18 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | The crystal structure of a Cupin 2 conserved barrel domain protein from Salinispora arenicola CNS-205
To be Published
|
|
4RXI
 
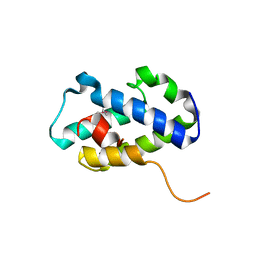 | | Structure of C-terminal domain of uncharacterized protein from Legionella pneumophila | | Descriptor: | hypothetical protein lpg0944 | | Authors: | Cuff, M, Nocek, B, Evdokimova, E, Egorova, O, Joachimiak, A, Ensminger, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-11 | | Release date: | 2015-05-06 | | Last modified: | 2017-01-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol Syst Biol, 12, 2016
|
|
4RSH
 
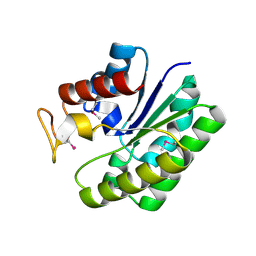 | | Structure of a putative lipolytic protein of G-D-S-L family from Desulfitobacterium hafniense DCB-2 | | Descriptor: | CHLORIDE ION, Lipolytic protein G-D-S-L family | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-07 | | Release date: | 2014-11-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of a putative lipolytic protein of G-D-S-L family from Desulfitobacterium hafniense DCB-2
To be Published
|
|
4RXV
 
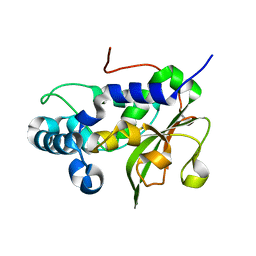 | | The crystal structure of the N-terminal fragment of uncharacterized protein from Legionella pneumophila | | Descriptor: | hypothetical protein lpg0944 | | Authors: | Nocek, B, Cuff, M, Evdokimova, E, Egorova, O, Joachimiak, A, Ensminger, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-12 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol Syst Biol, 12, 2016
|
|
4R0J
 
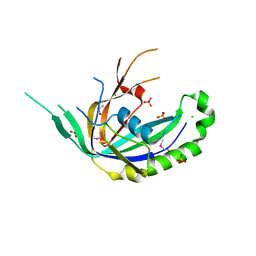 | | The crystal structure of a functionally uncharacterized protein SMU1763c from Streptococcus mutans | | Descriptor: | CHLORIDE ION, SULFATE ION, Uncharacterized protein | | Authors: | Tan, K, Xu, X, Cui, H, Liu, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-31 | | Release date: | 2014-08-13 | | Method: | X-RAY DIFFRACTION (1.715 Å) | | Cite: | The crystal structure of a functionally uncharacterized protein SMU1763c from Streptococcus mutans
To be Published
|
|
4RD8
 
 | | The crystal structure of a functionally-unknown protein from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | Uncharacterized protein | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-18 | | Release date: | 2014-10-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The crystal structure of a functionally-unknown protein from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To be Published
|
|
4R23
 
 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with dicloxacillin | | Descriptor: | (3R,4R,5R)-3-(2,6-dichlorophenyl)-N-{(1R)-1-[(2R,4S)-4-(dihydroxymethyl)-5,5-dimethyl-1,3-thiazolidin-2-yl]-2-oxoethyl} -5-methyl-1,2-oxazolidine-4-carboxamide, 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-08 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with dicloxacillin
To be Published
|
|
4RDC
 
 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with proline | | Descriptor: | Amino acid/amide ABC transporter substrate-binding protein, HAAT family, FORMIC ACID, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-18 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with proline.
To be Published
|
|
4RGR
 
 | | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, GLYCEROL, Repressor protein, ... | | Authors: | Kim, Y, Joachimiak, G, Bigelow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2015-03-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP
To be Published, 2014
|
|
4RGS
 
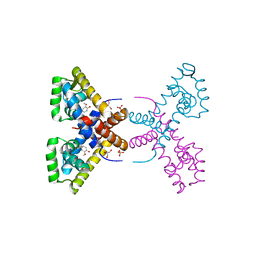 | | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with Vanilin | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, GLYCEROL, Repressor protein, ... | | Authors: | Kim, Y, Joachimiak, G, Bigelow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2015-03-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with Vanilin
To be Published, 2014
|
|
4RGU
 
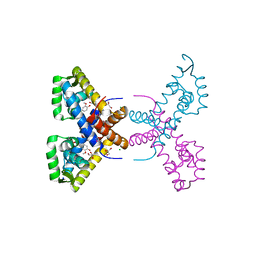 | | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with ferulic acid | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Joachimiak, G, Bigelow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2015-03-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with ferulic acid
To be Published, 2014
|
|
4RGX
 
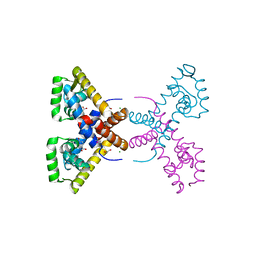 | | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with 3,4-dihydroxy bezoic acid | | Descriptor: | 1,2-ETHANEDIOL, 3,4-DIHYDROXYBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Joachimiak, G, Bigelow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2015-03-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with 3,4-dihydroxy bezoic acid
To be Published, 2014
|
|
