7STQ
 
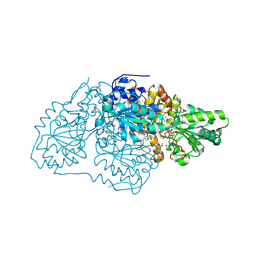 | | Crystal structure of arabidopsis thaliana acetohydroxyacid synthase W574L mutant in complex with chlorimuron-ethyl | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ... | | Authors: | Guddat, L.W, Cheng, Y. | | Deposit date: | 2021-11-15 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of resistance to herbicides that target acetohydroxyacid synthase.
Nat Commun, 13, 2022
|
|
7OFL
 
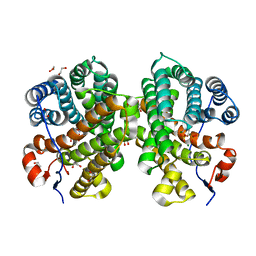 | | Crystal structure of the sesquiterpene synthase Copu9 from coniophora puteana in complex with alendronate | | Descriptor: | 1,2-ETHANEDIOL, 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE, MAGNESIUM ION, ... | | Authors: | Dimos, N, Loll, B. | | Deposit date: | 2021-05-05 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biotechnological potential and initial characterization of two novel sesquiterpene synthases from Basidiomycota Coniophora puteana for heterologous production of delta-cadinol.
Microb Cell Fact, 21, 2022
|
|
7U1D
 
 | |
7U25
 
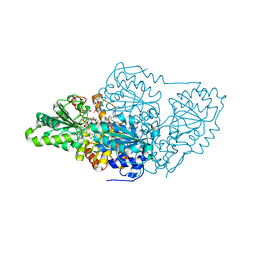 | | Crystal structure of arabidopsis thaliana acetohydroxyacid synthase W574L mutant in complex with bispyribac-sodium | | Descriptor: | 2,6-bis[(4,6-dimethoxypyrimidin-2-yl)oxy]benzoic acid, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ... | | Authors: | Guddat, L.W, Cheng, Y. | | Deposit date: | 2022-02-23 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural basis of resistance to herbicides that target acetohydroxyacid synthase.
Nat Commun, 13, 2022
|
|
7U1U
 
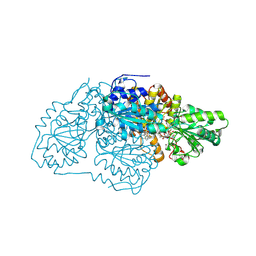 | |
7TZZ
 
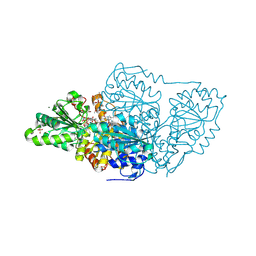 | | Crystal structure of arabidopsis thaliana acetohydroxyacid synthase P197T mutant in complex with bispyribac-sodium | | Descriptor: | 2,6-bis[(4,6-dimethoxypyrimidin-2-yl)oxy]benzoic acid, 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-2-[(1~{S})-1-(dioxidanyl)-1-oxidanyl-ethyl]-4-methyl-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Guddat, L.W, Cheng, Y. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis of resistance to herbicides that target acetohydroxyacid synthase.
Nat Commun, 13, 2022
|
|
6NZV
 
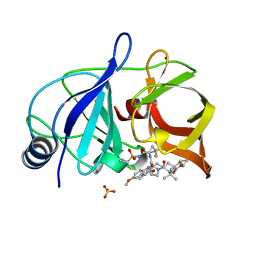 | | Crystal structure of HCV NS3/4A protease in complex with compound 12 | | Descriptor: | (1aR,5S,8S,9S,10R,22aR)-5-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-9-ethyl-14-methoxy-3,6-dioxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadecino[11,12-b]quinoxaline-8-carboxamide, HCV NS3/4A protease, SULFATE ION, ... | | Authors: | Appleby, T.C, Taylor, J.G. | | Deposit date: | 2019-02-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of the pan-genotypic hepatitis C virus NS3/4A protease inhibitor voxilaprevir (GS-9857): A component of Vosevi®.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
5GZR
 
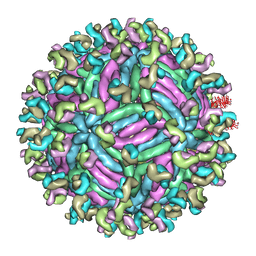 | | Zika virus E protein complexed with a neutralizing antibody Z23-Fab | | Descriptor: | Z23 Fab heavy chain, Z23 Fab light chain, structural protein E, ... | | Authors: | Gao, G.G, Shi, Y, Peng, R, Liu, S. | | Deposit date: | 2016-10-01 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
6G46
 
 | |
6INI
 
 | | a glycosyltransferase complex with UDP and the product | | Descriptor: | (8alpha,9beta,10alpha,13alpha)-13-{[beta-D-glucopyranosyl-(1->2)-[beta-D-glucopyranosyl-(1->3)]-beta-D-glucopyranosyl]oxy}kaur-16-en-18-oic acid, 1-O-[(8alpha,9beta,10alpha,13alpha)-13-(beta-D-glucopyranosyloxy)-18-oxokaur-16-en-18-yl]-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zhu, X, Yang, T, Naismith, J.H. | | Deposit date: | 2018-10-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
4GHU
 
 | | Crystal structure of TRAF3/Cardif | | Descriptor: | Mitochondrial antiviral-signaling protein, TNF receptor-associated factor 3 | | Authors: | Zhang, P. | | Deposit date: | 2012-08-08 | | Release date: | 2012-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Single Amino Acid Substitutions Confer the Antiviral Activity of the TRAF3 Adaptor Protein onto TRAF5
Sci.Signal., 5, 2012
|
|
2GKW
 
 | | Key contacts promote recongnito of BAFF-R by TRAF3 | | Descriptor: | TNF receptor-associated factor 3, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Ely, K.R. | | Deposit date: | 2006-04-03 | | Release date: | 2006-04-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Key molecular contacts promote recognition of the BAFF receptor by TNF receptor-associated factor 3: implications for intracellular signaling regulation.
J.Immunol., 173, 2004
|
|
1LRA
 
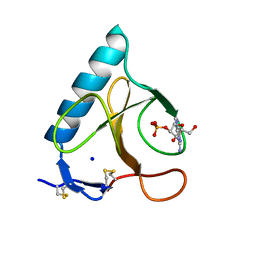 | | CRYSTALLOGRAPHIC STUDY OF GLU 58 ALA RNASE T1(ASTERISK)2'-GUANOSINE MONOPHOSPHATE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1, SODIUM ION | | Authors: | Pletinckx, J, Steyaert, J, Choe, H.-W, Heinemann, U, Wyns, L. | | Deposit date: | 1993-10-01 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic study of Glu58Ala RNase T1 x 2'-guanosine monophosphate at 1.9-A resolution.
Biochemistry, 33, 1994
|
|
6NZT
 
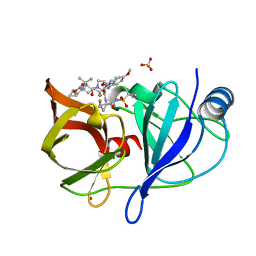 | |
6EG3
 
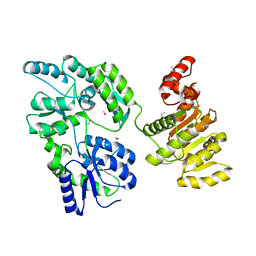 | | Crystal structure of human BRM in complex with compound 15 | | Descriptor: | 3-[(4-{[(2-chloropyridin-4-yl)carbamoyl]amino}pyridin-2-yl)ethynyl]benzoic acid, ETHANOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2 | | Authors: | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
6EG2
 
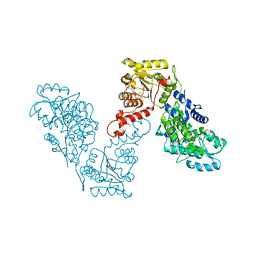 | | Crystal structure of human BRM in complex with compound 16 | | Descriptor: | ISOPROPYL ALCOHOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2, N-(5-amino-2-chloropyridin-4-yl)-N'-(4-bromo-3-{[3-(hydroxymethyl)phenyl]ethynyl}-1,2-thiazol-5-yl)urea | | Authors: | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
5H1Y
 
 | |
1RNT
 
 | | RESTRAINED LEAST-SQUARES REFINEMENT OF THE CRYSTAL STRUCTURE OF THE RIBONUCLEASE T1(ASTERISK)2(PRIME)-GUANYLIC ACID COMPLEX AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1 ISOZYME | | Authors: | Saenger, W, Arni, R, Heinemann, U, Tokuoka, R. | | Deposit date: | 1987-07-10 | | Release date: | 1987-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Restrained Least-Squares Refinement of the Crystal Structure of the Ribonuclease T1(Asterisk)2(Prime)-Guanylic Acid Complex at 1.9 Angstroms Resolution
Acta Crystallogr.,Sect.B, 43, 1987
|
|
1GOY
 
 | | HYDROLASE(ENDORIBONUCLEASE)RIBONUCLEASE BI(G SPECIFIC ENDONUCLEASE) (E.C.3.1.27.-) COMPLEXED WITH GUANOSINE-3'-PHOSPHATE (3'-GMP) | | Descriptor: | GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE, SULFATE ION | | Authors: | Polyakov, K.M, Lebedev, A.A, Pavlovsky, A.G, Sanishvili, R.G, Dodson, G.G. | | Deposit date: | 2001-10-26 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of Substrate-Free Microbial Ribonuclease Binase and of its Complexes with 3'Gmp and Sulfate Ions
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GOV
 
 | | RIBONUCLEASE BI(G SPECIFIC ENDONUCLEASE) COMPLEXED WITH SULFATE IONS | | Descriptor: | RIBONUCLEASE, SULFATE ION | | Authors: | Polyakov, K.M, Lebedev, A.A, Pavlovsky, A.G, Sanishvili, R.G, Dodson, G.G. | | Deposit date: | 2001-10-26 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of Substrate-Free Microbial Ribonuclease Binase and of its Complexes with 3'Gmp and Sulfate Ions
Acta Crystallogr.,Sect.D, 58, 2002
|
|
8EJ0
 
 | | Crystal structure of Fe-S cluster-dependent dehydratase from Paralcaligenes ureilyticus in complex with Mg | | Descriptor: | BICARBONATE ION, CARBON DIOXIDE, Dihydroxyacid dehydratase, ... | | Authors: | Bayaraa, T, Lonhienne, T, Guddat, L.W. | | Deposit date: | 2022-09-16 | | Release date: | 2023-08-30 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural and Functional Insight into the Mechanism of the Fe-S Cluster-Dependent Dehydratase from Paralcaligenes ureilyticus.
Chemistry, 29, 2023
|
|
9BAG
 
 | | Crystal structure of Oryza sativa ketol-acid reductoisomerase in complex with Mg2+, and JK-5-114 | | Descriptor: | 6-hydroxy-2-phenyl[1,3]thiazolo[4,5-d]pyrimidine-5,7(4H,6H)-dione, Ketol-acid reductoisomerase, chloroplastic, ... | | Authors: | Lin, X, Guddat, L.W. | | Deposit date: | 2024-04-04 | | Release date: | 2025-04-09 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A Ketol-Acid Reductoisomerase Inhibitor That Has Antituberculosis and Herbicidal Activity.
Chemistry, 31, 2025
|
|
9BAE
 
 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase in complex with Mg2+, and JK-5-115 | | Descriptor: | 6-hydroxy-2-phenyl[1,3]thiazolo[5,4-d]pyrimidine-5,7(4H,6H)-dione, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION | | Authors: | Kurz, J.L, Lin, X, Guddat, L.W. | | Deposit date: | 2024-04-03 | | Release date: | 2025-04-09 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | A Ketol-Acid Reductoisomerase Inhibitor That Has Antituberculosis and Herbicidal Activity.
Chemistry, 31, 2025
|
|
8CY8
 
 | |
2ZOA
 
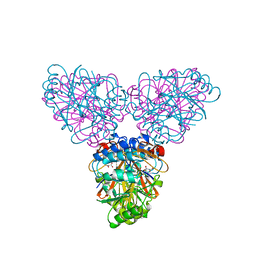 | | Malonate-bound structure of the glycerophosphodiesterase from Enterobacter aerogenes (GpdQ) COLLECTED AT 1.280 ANGSTROM | | Descriptor: | FE (II) ION, MALONATE ION, Phosphohydrolase | | Authors: | Ollis, D.L, Jackson, C.J, Carr, P.D. | | Deposit date: | 2008-05-07 | | Release date: | 2008-10-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Malonate-bound structure of the glycerophosphodiesterase from Enterobacter aerogenes (GpdQ) and characterization of the native Fe2+ metal-ion preference.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
