4V5A
 
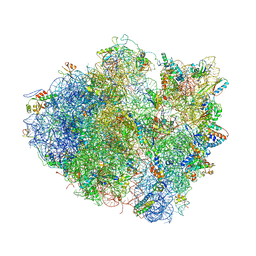 | | Structure of the Ribosome Recycling Factor bound to the Thermus thermophilus 70S ribosome with mRNA, ASL-Phe and tRNA-fMet | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Weixlbaumer, A, Petry, S, Dunham, C.M, Selmer, M, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2007-06-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the ribosome recycling factor bound to the ribosome.
Nat. Struct. Mol. Biol., 14, 2007
|
|
4USX
 
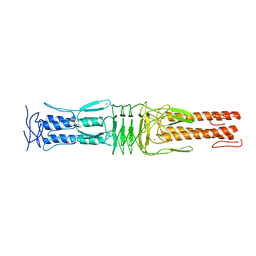 | | The Structure of the C-terminal YadA-like domain of BPSL2063 from Burkholderia pseudomallei | | Descriptor: | MAGNESIUM ION, TRIMERIC AUTOTRANSPORTER ADHESIN | | Authors: | Perletti, L, Gourlay, L.J, Peano, C, Pietrelli, A, DeBellis, G, Deantonio, C, Santoro, C, Sblattero, D, Bolognesi, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selecting Soluble/Foldable Protein Domains Through Single-Gene or Genomic Orf Filtering: Structure of the Head Domain of Burkholderia Pseudomallei Antigen Bpsl2063.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UYW
 
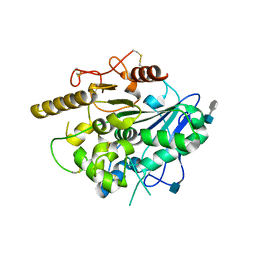 | |
4UZI
 
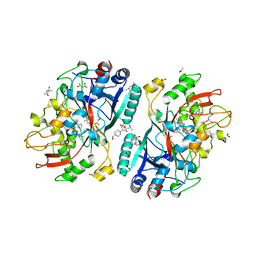 | | Crystal Structure of AauDyP Complexed with Imidazole | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Strittmatter, E, Liers, C, Ullrich, R, Hofrichter, M, Piontek, K, Plattner, D.A. | | Deposit date: | 2014-09-05 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Toolbox of Auricularia Auricula-Judae Dye-Decolorizing Peroxidase - Identification of Three New Potential Substrate-Interaction Sites.
Arch.Biochem.Biophys., 574, 2015
|
|
4V4O
 
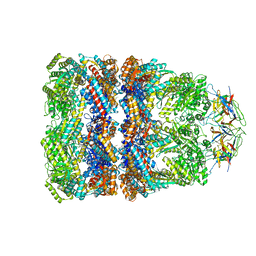 | | Crystal Structure of the Chaperonin Complex Cpn60/Cpn10/(ADP)7 from Thermus Thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Shimamura, T, Koike-Takeshita, A, Yokoyama, K, Masui, R, Murai, N, Yoshida, M, Taguchi, H, Iwata, S. | | Deposit date: | 2004-05-23 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the native chaperonin complex from Thermus thermophilus revealed unexpected asymmetry at the cis-cavity
STRUCTURE, 12, 2004
|
|
4V87
 
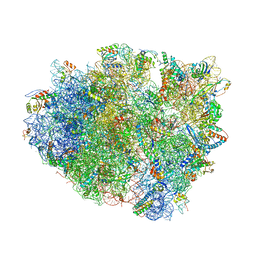 | | Crystal structure analysis of ribosomal decoding. | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Demeshkina, N, Jenner, L, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-09-20 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4V8P
 
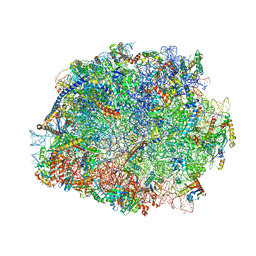 | | T.thermophila 60S ribosomal subunit in complex with initiation factor 6. | | Descriptor: | 26S RRNA, 5.8S RRNA, 5S RRNA, ... | | Authors: | Klinge, S, Voigts-Hoffmann, F, Leibundgut, M, Arpagaus, S, Ban, N. | | Deposit date: | 2011-09-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Crystal Structure of the Eukaryotic 60S Ribosomal Subunit in Complex with Initiation Factor 6.
Science, 334, 2011
|
|
7ZUF
 
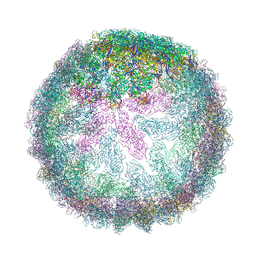 | | Saccharomyces cerevisiae L-BC virus, open particle, C5 reconstruction | | Descriptor: | Major capsid protein | | Authors: | Grybchuk, D, Prochazkova, M, Fuzik, T, Konovalovas, A, Serva, S, Yurchenko, V, Plevka, P. | | Deposit date: | 2022-05-12 | | Release date: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structures of L-BC virus and its open particle provide insight into Totivirus capsid assembly.
Commun Biol, 5, 2022
|
|
8A3O
 
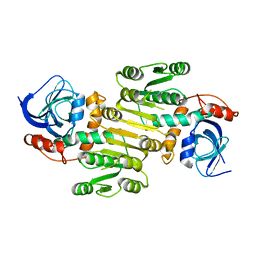 | | Structure of human Fy-4 | | Descriptor: | Quinone oxidoreductase-like protein 1 | | Authors: | Schuhmacher, J.S, Zerial, M. | | Deposit date: | 2022-06-08 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of mRNA binding by the human FERRY Rab5 effector complex.
Mol.Cell, 83, 2023
|
|
8A3P
 
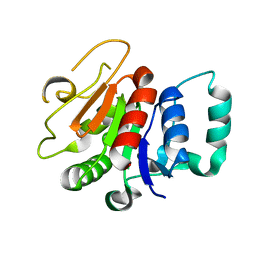 | | Structure of human Fy-5. | | Descriptor: | Glutamine amidotransferase-like class 1 domain-containing protein 1 | | Authors: | Schuhmacher, J.S, Zerial, M. | | Deposit date: | 2022-06-08 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of mRNA binding by the human FERRY Rab5 effector complex.
Mol.Cell, 83, 2023
|
|
4V5O
 
 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC 40S RIBOSOMAL SUBUNIT IN COMPLEX WITH INITIATION FACTOR 1. | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S12, 40S RIBOSOMAL PROTEIN S3A, ... | | Authors: | Rabl, J, Leibundgut, M, Ataide, S.F, Haag, A, Ban, N. | | Deposit date: | 2010-11-26 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.93 Å) | | Cite: | Crystal Structure of the Eukaryotic 40S Ribosomal Subunit in Complex with Initiation Factor 1.
Science, 331, 2011
|
|
4V63
 
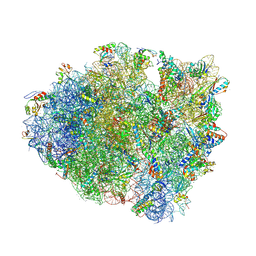 | | Structural basis for translation termination on the 70S ribosome. | | Descriptor: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | Authors: | Laurberg, M, Asahara, H, Korostelev, A, Zhu, J, Trakhanov, S, Noller, H.F. | | Deposit date: | 2008-05-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.207 Å) | | Cite: | Structural basis for translation termination on the 70S ribosome
Nature, 454, 2008
|
|
4V5V
 
 | | Structure of respiratory syncytial virus nucleocapsid protein, P1 crystal form | | Descriptor: | RESPIRATORY SYNCYTIAL VIRUS NUCLEOCAPSID PROTEIN, RNA | | Authors: | El Omari, K, Dhaliwal, B, Ren, J, Abrescia, N.G.A, Lockyer, M, Powell, K.L, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2011-05-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of Respiratory Syncytial Virus Nucleocapsid Protein from Two Crystal Forms: Details of Potential Packing Interactions in the Native Helical Form.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4US1
 
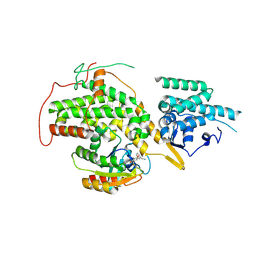 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | (3S)-3-[3-(aminomethyl)phenyl]-1-ethylpyrrolidine-2,5-dione, GTPASE HRAS, SON OF SEVENLESS HOMOLOG 1 | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
4USZ
 
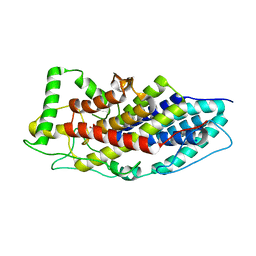 | | Crystal structure of the first bacterial vanadium dependant iodoperoxidase | | Descriptor: | SODIUM ION, VANADATE ION, VANADIUM-DEPENDENT HALOPEROXIDASE | | Authors: | Rebuffet, E, Delage, L, Fournier, J.B, Rzonca, J, Potin, P, Michel, G, Czjzek, M, Leblanc, C. | | Deposit date: | 2014-07-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Bacterial Vanadium Iodoperoxidase from the Marine Flavobacteriaceae Zobellia Galactanivorans Reveals Novel Molecular and Evolutionary Features of Halide Specificity in This Enzyme Family.
Appl.Environ.Microbiol., 80, 2014
|
|
7ZOS
 
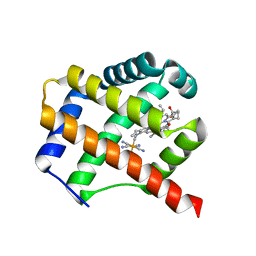 | | Class 1 Phytoglobin from Sugar beet (BvPgb1.2) | | Descriptor: | CYANIDE ION, HEXACYANOFERRATE(3-), Non-symbiotic hemoglobin class 1, ... | | Authors: | Nyblom, M, Christensen, S, Eriksson, N, Bulow, L. | | Deposit date: | 2022-04-26 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Oxidative Implications of Substituting a Conserved Cysteine Residue in Sugar Beet Phytoglobin BvPgb 1.2.
Antioxidants, 11, 2022
|
|
4V16
 
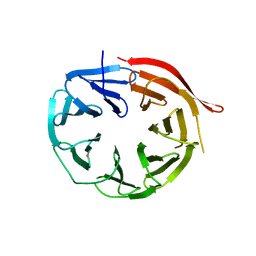 | | KlHsv2 with loop 6CD replaced by a Gly-Ser linker | | Descriptor: | SVP1-LIKE PROTEIN 2 | | Authors: | Busse, R.A, Scacioc, A, Krick, R, Perez-Lara, A, Thumm, M, Kuhnel, K. | | Deposit date: | 2014-09-25 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of Proppin-Phosphoinositide Binding and Role of Loop 6Cd in Proppin-Membrane Binding.
Biophys.J., 108, 2015
|
|
4UXN
 
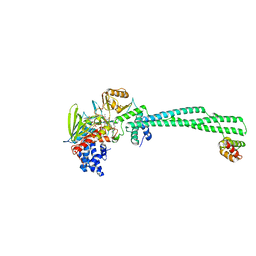 | | LSD1(KDM1A)-CoREST in complex with Z-Pro derivative of MC2580 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, ... | | Authors: | Rodriguez, V, Valente, S, Stazi, G, Lucidi, A, Mercurio, C, Vianello, P, Ciossani, G, Mattevi, A, Botrugno, O.A, Dessanti, P, Minucci, S, Varasi, M, Mai, A. | | Deposit date: | 2014-08-27 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Pyrrole- and Indole-Containing Tranylcypromine Derivatives as Novel Lysine-Specific Demethylase 1 Inhibitors Active on Cancer Cells
Chemmedchem, 6, 2015
|
|
4UY4
 
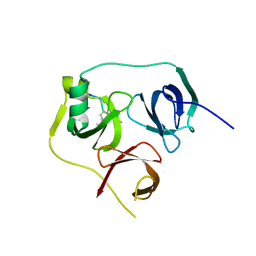 | | 1.86 A structure of human Spindlin-4 protein in complex with histone H3K4me3 peptide | | Descriptor: | GLYCEROL, HISTONE H3K4ME3, SPINDLIN-4 | | Authors: | Talon, R, Gileadi, C, Johansson, C, Burgess-Brown, N, Shrestha, L, von Delft, F, Krojer, T, Fairhead, M, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U. | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | 1.86 A Structure of Human Spindlin-4 Protein in Complex with Histone H3K4Me3 Peptide
To be Published
|
|
4UYU
 
 | |
4V19
 
 | | Structure of the large subunit of the mammalian mitoribosome, part 1 of 2 | | Descriptor: | MAGNESIUM ION, MITORIBOSOMAL 16S RRNA, MITORIBOSOMAL CP TRNA, ... | | Authors: | Greber, B.J, Boehringer, D, Leibundgut, M, Bieri, P, Leitner, A, Schmitz, N, Aebersold, R, Ban, N. | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The Complete Structure of the Large Subunit of the Mammalian Mitochondrial Ribosome
Nature, 515, 2014
|
|
4V51
 
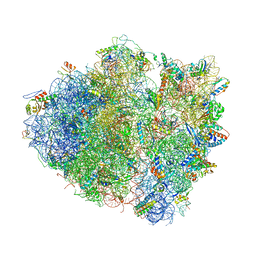 | | Structure of the Thermus thermophilus 70S ribosome complexed with mRNA, tRNA and paromomycin | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Selmer, M, Dunham, C.M, Murphy, F.V, Weixlbaumer, A, Petry, S, Weir, J.R, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2006-07-31 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the 70S ribosome complexed with mRNA and tRNA.
Science, 313, 2006
|
|
4V8C
 
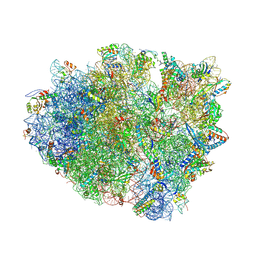 | | Crystal structure analysis of ribosomal decoding (near-cognate tRNA-leu complex with paromomycin). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4V67
 
 | | Crystal structure of a translation termination complex formed with release factor RF2. | | Descriptor: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Asahara, H, Lancaster, L, Laurberg, M, Hirschi, A, Noller, H.F. | | Deposit date: | 2008-10-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a translation termination complex formed with release factor RF2.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4V6T
 
 | | Structure of the bacterial ribosome complexed by tmRNA-SmpB and EF-G during translocation and MLD-loading | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Ramrath, D.J.F, Yamamoto, H, Rother, K, Wittek, D, Pech, M, Mielke, T, Loerke, J, Scheerer, P, Ivanov, P, Teraoka, Y, Shpanchenko, O, Nierhaus, K.H, Spahn, C.M.T. | | Deposit date: | 2012-01-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | The complex of tmRNA-SmpB and EF-G on translocating ribosomes.
Nature, 485, 2012
|
|
