7E9P
 
 | |
7E9N
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(1 down RBD, state1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-04-06 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7ENF
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with Fab30 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab30, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7ENG
 
 | |
7F46
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab (state1, local refinement of the RBD, NTD and 35B5 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-06-17 | | Release date: | 2022-03-23 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (4.79 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7YEP
 
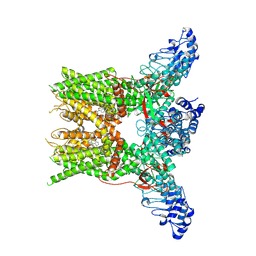 | | 2-APB bound state of mTRPV2 | | Descriptor: | 2-aminoethyl diphenylborinate, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-07-06 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
8H77
 
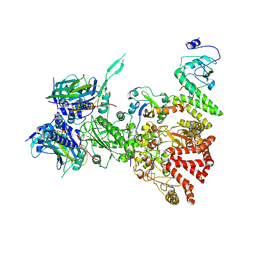 | | Hsp90-AhR-p23-XAP2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, Aryl hydrocarbon receptor, ... | | Authors: | Wen, Z.L, Zhai, Y.J, Zhu, Y, Sun, F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the cytosolic AhR complex.
Structure, 31, 2023
|
|
7XOZ
 
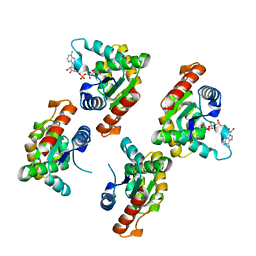 | | Crystal structure of RPPT-TIR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase | | Authors: | Song, W, Jia, A, Huang, S, Chai, J. | | Deposit date: | 2022-05-02 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | TIR-catalyzed ADP-ribosylation reactions produce signaling molecules for plant immunity.
Science, 377, 2022
|
|
3E6F
 
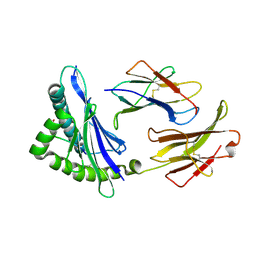 | | MHC CLASS I H-2Dd Heavy chain complexed with Beta-2 Microglobulin and a variant peptide, PA9, from the Human immunodeficiency virus (BaL) envelope glycoprotein 120 | | Descriptor: | BETA-2 MICROGLOBULIN, Envelope glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Wang, R, Natarajan, K, Robinson, H, Margulies, D.H. | | Deposit date: | 2008-08-15 | | Release date: | 2009-08-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Different vaccine vectors delivering the same antigen elicit CD8+ T cell responses with distinct clonotype and epitope specificity
J.Immunol., 183, 2009
|
|
3E6H
 
 | | MHC CLASS I H-2Dd heavy chain complexed with Beta-2 Microglobulin and a variant peptide, PI10, from the human immunodeficiency virus (BaL) envelope glycoprotein 120 | | Descriptor: | Envelope glycoprotein 10-residue peptide, H-2 class I histocompatibility antigen, D-D alpha chain, ... | | Authors: | Wang, R, Natarajan, K, Robinson, H, Margulies, D.H. | | Deposit date: | 2008-08-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Different vaccine vectors delivering the same antigen elicit CD8+ T cell responses with distinct clonotype and epitope specificity
J.Immunol., 183, 2009
|
|
3R24
 
 | | Crystal structure of nsp10/nsp16 complex of SARS coronavirus | | Descriptor: | 2'-O-methyl transferase, Non-structural protein 10 and Non-structural protein 11, S-ADENOSYLMETHIONINE, ... | | Authors: | Liu, X, Guo, D, Su, C, Chen, Y. | | Deposit date: | 2011-03-13 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex.
Plos Pathog., 7, 2011
|
|
2K6B
 
 | |
2M80
 
 | | Solution structure of yeast dithiol glutaredoxin Grx8 | | Descriptor: | Glutaredoxin-8 | | Authors: | Tang, Y, Zhang, J, Yu, J, Wu, J, Zhou, C.Z, Shi, Y. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-guided activity enhancement and catalytic mechanism of yeast grx8
Biochemistry, 53, 2014
|
|
7VRG
 
 | | Crystal structure of chitinase-h from O. furnacalis in complex with Lynamicin B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, ... | | Authors: | Lu, Q, Liu, T, Zhou, Y, Yang, Q. | | Deposit date: | 2021-10-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lynamicin B is a Potential Pesticide by Acting as a Lepidoptera-Exclusive Chitinase Inhibitor.
J.Agric.Food Chem., 69, 2021
|
|
4I3R
 
 | | Crystal structure of the outer domain of HIV-1 gp120 in complex with VRC-PG04 space group P3221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of VRC-PG04 Fab, Light chain of VRC-PG04 Fab, ... | | Authors: | Joyce, M.G, Biertumpfel, C, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2012-11-26 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Outer Domain of HIV-1 gp120: Antigenic Optimization, Structural Malleability, and Crystal Structure with Antibody VRC-PG04.
J.Virol., 87, 2013
|
|
4I3S
 
 | | Crystal structure of the outer domain of HIV-1 gp120 in complex with VRC-PG04 space group P21 | | Descriptor: | CALCIUM ION, Heavy chain of VRC-PG04 Fab, Light chain of VRC-PG04 Fab, ... | | Authors: | Joyce, M.G, Biertumpfel, C, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2012-11-26 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Outer Domain of HIV-1 gp120: Antigenic Optimization, Structural Malleability, and Crystal Structure with Antibody VRC-PG04.
J.Virol., 87, 2013
|
|
7C7P
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, CHLORIDE ION | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Yang, S.Y, Lei, J. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
7D3I
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with MI-23 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[3-[3,5-bis(fluoranyl)phenyl]propanoyl]-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase | | Authors: | Zeng, R, Li, Y.S, Qiao, J.X, Wang, Y.F, Yang, S.Y, Lei, J. | | Deposit date: | 2020-09-19 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
7COM
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Boceprevir (space group P212121) | | Descriptor: | 3C-like proteinase, boceprevir (bound form) | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Liu, J.M, Zhou, Y.L, Chen, P, Yang, S.Y, Lei, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
5XH3
 
 | | Crystal structure of a novel PET hydrolase R103G/S131A mutant in complex with HEMT from Ideonella sakaiensis 201-F6 | | Descriptor: | GLYCEROL, O 4-(2-hydroxyethyl) O 1-methyl benzene-1,4-dicarboxylate, Poly(ethylene terephthalate) hydrolase, ... | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
5Y9P
 
 | | Staphylococcus aureus RNase HII | | Descriptor: | GLYCEROL, Ribonuclease HII | | Authors: | Hang, T, Wu, M, Zhang, X. | | Deposit date: | 2017-08-26 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into a novel functional dimer of Staphylococcus aureus RNase HII
Biochem. Biophys. Res. Commun., 503, 2018
|
|
6A5A
 
 | |
5Z8Q
 
 | |
5Z8I
 
 | |
6A5D
 
 | |
