8VB1
 
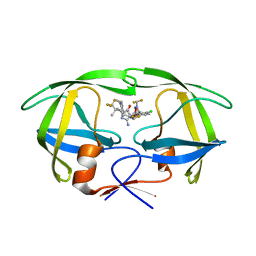 | | Crystal structure of HIV-1 protease with GS-9770 | | Descriptor: | (2S)-2-{(3M)-4-chloro-3-[1-(difluoromethyl)-1H-1,2,4-triazol-5-yl]phenyl}-2-[(2E,4R)-4-[4-(2-cyclopropyl-2H-1,2,3-triazol-4-yl)phenyl]-2-imino-5-oxo-4-(3,3,3-trifluoro-2,2-dimethylpropyl)imidazolidin-1-yl]ethyl [1-(difluoromethyl)cyclopropyl]carbamate, HIV-1 protease | | Authors: | Lansdon, E.B. | | Deposit date: | 2023-12-11 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Preclinical characterization of a non-peptidomimetic HIV protease inhibitor with improved metabolic stability.
Antimicrob.Agents Chemother., 68, 2024
|
|
4XPM
 
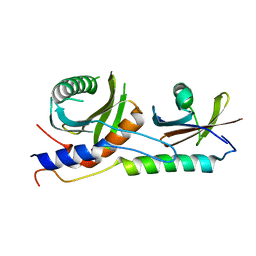 | | Crystal structure of EGO-TC | | Descriptor: | Protein MEH1, Protein SLM4, Uncharacterized protein YCR075W-A | | Authors: | Powis, K, Zhang, T, De Virgilio, C, Ding, J. | | Deposit date: | 2015-01-17 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the Ego1-Ego2-Ego3 complex and its role in promoting Rag GTPase-dependent TORC1 signaling.
Cell Res., 25, 2015
|
|
8UWR
 
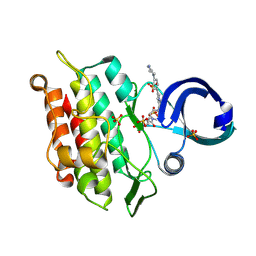 | | Crystal structure of human ACVR1 (ALK2) kinase in complex with compound 3 | | Descriptor: | Activin receptor type-1, SULFATE ION, cyclopropyl(4-{(8R)-6-[4-(piperazin-1-yl)phenyl]pyrrolo[1,2-b]pyridazin-4-yl}piperazin-1-yl)methanone | | Authors: | Kim, J.L. | | Deposit date: | 2023-11-07 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | An ALK2 inhibitor, BLU-782, prevents heterotopic ossification in a mouse model of fibrodysplasia ossificans progressiva.
Sci Transl Med, 16, 2024
|
|
8UWN
 
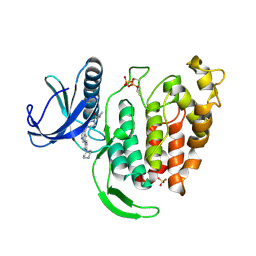 | | Crystal structure of human CLK1 kinase in complex with compound 3 | | Descriptor: | Dual specificity protein kinase CLK1, SULFATE ION, cyclopropyl(4-{(8R)-6-[4-(piperazin-1-yl)phenyl]pyrrolo[1,2-b]pyridazin-4-yl}piperazin-1-yl)methanone | | Authors: | Kim, J.L. | | Deposit date: | 2023-11-07 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An ALK2 inhibitor, BLU-782, prevents heterotopic ossification in a mouse model of fibrodysplasia ossificans progressiva.
Sci Transl Med, 16, 2024
|
|
4YH3
 
 | | Crystal structure of human BRD4(1) in complex with 4-[(2E)-3-(4-methoxyphenyl)-2-phenylprop-2-enoyl]-3,4-dihydroquinoxalin-2(1H)-one (compound 19a) | | Descriptor: | 4-[(2E)-3-(4-methoxyphenyl)-2-phenylprop-2-enoyl]-3,4-dihydroquinoxalin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | White, A, Lakshminarasimhan, D, Suto, R.K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of a new chemical series of BRD4(1) inhibitors using protein-ligand docking and structure-guided design.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6WYV
 
 | | E. coli 50S ribosome bound to compounds 47 and VS1 | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2020-05-13 | | Release date: | 2020-06-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
4YH4
 
 | | Crystal structure of human BRD4(1) in complex with 4-[(5-phenylpyridin-3-yl)carbonyl]-3,4-dihydroquinoxalin-2(1H)-one (compound 19d) | | Descriptor: | 4-[(5-phenylpyridin-3-yl)carbonyl]-3,4-dihydroquinoxalin-2(1H)-one, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of a new chemical series of BRD4(1) inhibitors using protein-ligand docking and structure-guided design.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7X2M
 
 | | Crystal structure of nanobody 1-2C7 with SARS-CoV-2 RBD | | Descriptor: | 1-2C7, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2L
 
 | | Crystal structure of nanobody 3-2A2-4 with SARS-CoV-2 RBD | | Descriptor: | Nanobody 3-2A2-4, Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2J
 
 | | Crystal structure of nanobody Nb70 with SARS-CoV RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nb70, Spike protein S1 | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2K
 
 | | Crystal structure of nanobody Nb70 with antibody 1F11 fab and SARS-CoV-2 RBD | | Descriptor: | 1F11-H, 1F11-L, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7XGW
 
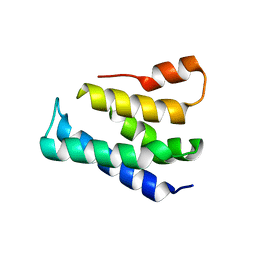 | |
8JGC
 
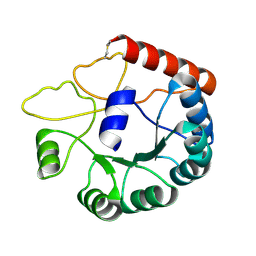 | | Cryo-EM structure of Mi3 fused with LOV2 | | Descriptor: | LOV domain-containing protein,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase | | Authors: | Zhang, H.W, Kang, W, Xue, C. | | Deposit date: | 2023-05-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Dynamic Metabolons Using Stimuli-Responsive Protein Cages.
J.Am.Chem.Soc., 146, 2024
|
|
8JGA
 
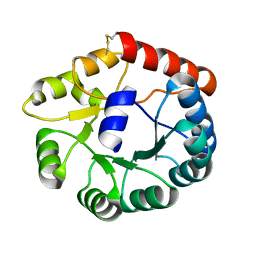 | | Cryo-EM structure of Mi3 fused with FKBP | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase | | Authors: | Zhang, H.W, Kang, W, Xue, C. | | Deposit date: | 2023-05-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Dynamic Metabolons Using Stimuli-Responsive Protein Cages.
J.Am.Chem.Soc., 146, 2024
|
|
8IKX
 
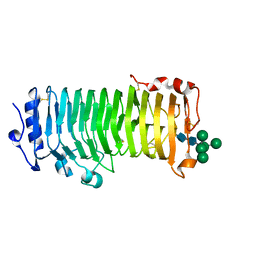 | | An Arabidopsis polygalacturonase PGLR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pectin lyase-like superfamily protein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Xiao, Y, Chai, J. | | Deposit date: | 2023-03-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A plant mechanism of hijacking pathogen virulence factors to trigger innate immunity.
Science, 383, 2024
|
|
8IKW
 
 | | A complex structure of PGIP-PG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Endo-polygalacturonase, ... | | Authors: | Xiao, Y, Chai, J. | | Deposit date: | 2023-03-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A plant mechanism of hijacking pathogen virulence factors to trigger innate immunity.
Science, 383, 2024
|
|
8JM9
 
 | |
8JMI
 
 | |
8JMA
 
 | |
8JME
 
 | |
8JMH
 
 | |
8DKE
 
 | |
8DKW
 
 | |
8DKI
 
 | |
8DKX
 
 | |
