1VKA
 
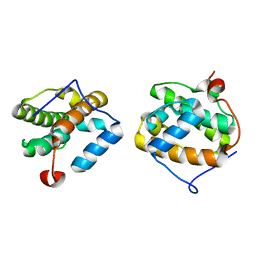 | | Southeast Collaboratory for Structural Genomics: Hypothetical Human Protein Q15691 N-Terminal Fragment | | Descriptor: | Microtubule-associated protein RP/EB family member 1 | | Authors: | Liu, Z.-J, Tempel, W, Schubot, F.D, Shah, A, Dailey, T.A, Mayer, M.R, Rose, J.P, Dailey, H.A, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-10 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Southeast Collaboratory for Structural Genomics: Hypothetical Human Protein Q15691 N-Terminal Fragment
TO BE PUBLISHED
|
|
1VJK
 
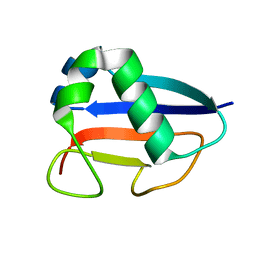 | | Putative molybdopterin converting factor, subunit 1 from Pyrococcus furiosus, Pfu-562899-001 | | Descriptor: | molybdopterin converting factor, subunit 1 | | Authors: | Chen, L, Liu, Z.J, Tempel, W, Shah, A, Lee, D, Rose, J.P, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Putative molybdopterin converting factor, subunit 1 from Pyrococcus furiosus, Pfu-562899-001 '
To be published
|
|
4R3H
 
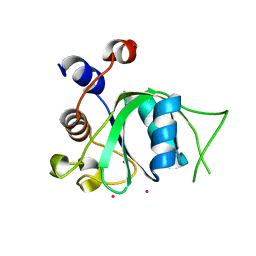 | | The crystal structure of an apo RNA binding protein | | Descriptor: | SULFATE ION, UNKNOWN ATOM OR ION, YTH domain-containing protein 1 | | Authors: | Xu, C, Liu, K, Tempel, W, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-08-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for selective binding of m(6)A RNA by the YTHDC1 YTH domain.
Nat.Chem.Biol., 10, 2014
|
|
4IH8
 
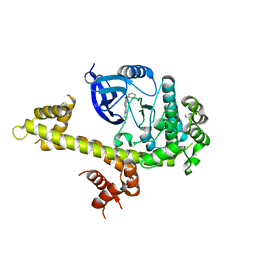 | | Crystal structure of TgCDPK1 with inhibitor bound | | Descriptor: | 4-Amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl-cyclopentane, Calmodulin-domain protein kinase 1 | | Authors: | El Bakkouri, M, Tempel, W, Crandall, I, Massad, T, Loppnau, P, Graslund, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kain, K, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.877 Å) | | Cite: | Crystal structure of TgCDPK1 with inhibitor bound
To be Published
|
|
1VKC
 
 | | Putative acetyl transferase from Pyrococcus furiosus | | Descriptor: | IODIDE ION, putative acetyl transferase | | Authors: | Habel, J.E, Liu, Z.-J, Tempel, W, Rose, J.P, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-11 | | Release date: | 2004-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Putative acetyl transferase from Pyrococcus furiosus
To be published
|
|
4IFG
 
 | | Crystal structure of TgCDPK1 with inhibitor bound | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, Calmodulin-domain protein kinase 1, UNKNOWN ATOM OR ION | | Authors: | El Bakkouri, M, Tempel, W, Crandall, I, Massad, T, Loppnau, P, Graslund, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kain, K, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-14 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of TgCDPK1 with inhibitor bound
To be Published
|
|
4RXX
 
 | | Crystal Structure of the N-terminal Domain of Human Ubiquitin Specific Protease 38 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Shen, L, Hu, J, Li, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-12 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of the N-terminal Domain of Human Ubiquitin Specific Protease 38
to be published
|
|
4RZ0
 
 | | Crystal Structure of Plasmodium falciparum putative histone methyltransferase PFL0690c | | Descriptor: | PFL0690c, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Ravichandran, M, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-17 | | Release date: | 2015-01-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Crystal Structure of Plasmodium falciparum putative histone methyltransferase PFL0690c
To be Published
|
|
4RZ7
 
 | | Crystal Structure of PVX_084705 with bound PCI32765 | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, UNKNOWN ATOM OR ION, cGMP-dependent protein kinase, ... | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, El Bakkouri, M, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Crystal Structure of PVX_084705 with bound PCI32765
To be Published
|
|
4PXW
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) | | Descriptor: | Protein VPRBP, UNKNOWN ATOM OR ION | | Authors: | Xu, C, Tempel, W, He, H, Li, Y, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of human DCAF1 WD40 repeats (Q1250L)
TO BE PUBLISHED
|
|
4ROF
 
 | | Crystal Structure of WW3 domain of ITCH in complex with TXNIP peptide | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Thioredoxin-interacting protein | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of WW3 domain of ITCH in complex with TXNIP peptide
To be Published
|
|
4Q94
 
 | | human RPRD1B CID in complex with a RPB1-CTD derived Ser2 phosphorylated peptide | | Descriptor: | Regulation of nuclear pre-mRNA domain-containing protein 1B, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Ni, Z, Xu, C, Tempel, W, El Bakkouri, M, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Greenblatt, J.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-04-29 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4QQI
 
 | | Crystal structure of ANKRA2-RFX7 complex | | Descriptor: | Ankyrin repeat family A protein 2, DNA-binding protein RFX7, SULFATE ION, ... | | Authors: | Xu, C, Tempel, W, Dong, A, Mackenzie, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of ANKRA2-RFX7 complex
To Be Published
|
|
4JXM
 
 | | Crystal structure of RRP9 WD40 repeats | | Descriptor: | U3 small nucleolar RNA-interacting protein 2, UNKNOWN ATOM OR ION | | Authors: | Wu, X, Tempel, W, Xu, C, El Bakkouri, M, He, H, Seitova, A, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-28 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of RRP9 WD40 repeats
To be Published
|
|
4QQ4
 
 | | CW-type zinc finger of MORC3 in complex with the amino terminus of histone H3 | | Descriptor: | CHLORIDE ION, Histone H3.3, MORC family CW-type zinc finger protein 3, ... | | Authors: | Liu, Y, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-26 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Family-wide Characterization of Histone Binding Abilities of Human CW Domain-containing Proteins.
J.Biol.Chem., 291, 2016
|
|
1YEM
 
 | | Conserved hypothetical protein Pfu-838710-001 from Pyrococcus furiosus | | Descriptor: | Conserved hypothetical protein Pfu-838710-001, PLATINUM (II) ION, UNKNOWN ATOM OR ION | | Authors: | Yang, H, Chang, J, Shah, A, Ng, J.D, Liu, Z.-J, Chen, L, Lee, D, Tempel, W, Praissman, J.L, Lin, D, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-28 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved hypothetical protein Pfu-838710-001 from Pyrococcus furiosus
To be published
|
|
4LD6
 
 | | PWWP domain of human PWWP Domain-Containing Protein 2B | | Descriptor: | GLYCEROL, PWWP domain-containing protein 2B, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Dombrovski, L, Tempel, W, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of PWWP domain of human PWWP Domain-Containing Protein 2B
To be Published
|
|
6BPI
 
 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragment peptide conjugates | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, MLY-SER-THR-E2G, ... | | Authors: | MADER, P, Mendoza-Sanchez, R, DONG, A, DOBROVETSKY, E, IQBAL, A, CORLESS, V, TEMPEL, W, LIEW, S.K, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D.B, SCHAPIRA, M, VEDADI, M, BROWN, P.J, Santhakumar, V, FRYE, S, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-23 | | Release date: | 2017-12-27 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain with aryl triazole fragment peptide conjugates
to be published
|
|
6D0S
 
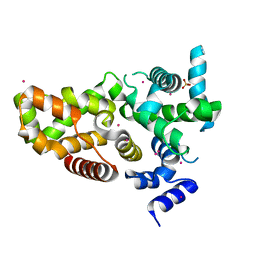 | | RabGAP domain of human TBC1D22B | | Descriptor: | SULFATE ION, TBC1 domain family member 22B, UNKNOWN ATOM OR ION | | Authors: | Tong, Y, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RabGAP domain of human TBC1D22B
To be Published
|
|
6CCT
 
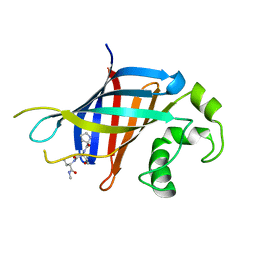 | | Fragment of GID4 in complex with a short peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Tetrapeptide | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
6CKO
 
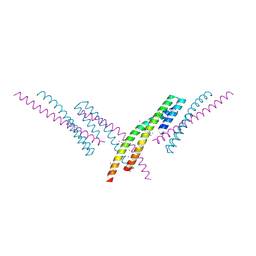 | | Crystal structure of an AF10 fragment | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, Protein AF-10, ... | | Authors: | Zhang, H, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the DOT1L-AF10 complex reveals mechanistic insights into MLL-AF10-associated leukemogenesis.
Genes Dev., 32, 2018
|
|
6D1T
 
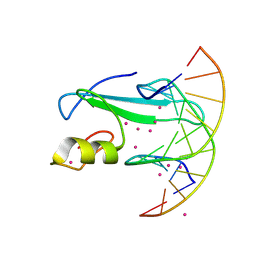 | | Complex of MBD1-MBD and methylated DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 1, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Xu, C, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-12 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Complex of MBD1-MBD and methylated DNA
to be published
|
|
6CD8
 
 | | Complex of GID4 fragment with short peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Tetrapeptide PSRV, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
6CDC
 
 | | GID4 in complex with a tetrapeptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Tetrapeptide PGLW, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
6CKN
 
 | | Crystal structure of an AF10 fragment | | Descriptor: | Protein AF-10, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural and functional analysis of the DOT1L-AF10 complex reveals mechanistic insights into MLL-AF10-associated leukemogenesis.
Genes Dev., 32, 2018
|
|
