8XFT
 
 | | LGR4-RSPO2-ZNRF3(1:1:1) | | Descriptor: | E3 ubiquitin-protein ligase ZNRF3, Leucine-rich repeat-containing G-protein coupled receptor 4, MB52, ... | | Authors: | Lu, W, Yong, G. | | Deposit date: | 2023-12-14 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural insights into the LGR4-RSPO2-ZNRF3 complexes regulating WNT/ beta-catenin signaling.
Nat Commun, 16, 2025
|
|
5UCM
 
 | |
8XTZ
 
 | | Crystal structure of Lsd18 in complex with a substrate | | Descriptor: | (4R,5S)-4-methyl-5-phenyl-3-((2R,3S,4S,6E,10E)-2,6,10-triethyl-3-hydroxy-4-methyldodeca-6,10-dienoyl)oxazolidin-2-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Deng, Y.M, Hu, Y.L, Chen, X. | | Deposit date: | 2024-01-12 | | Release date: | 2025-01-15 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Sequential Enantioselective Epoxidation by a Flavin-Dependent Monooxygenase in Lasalocid A Biosynthesis.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9CCV
 
 | | Crystal structure of human respiratory syncytial virus NS1 bound to human MED25 ACID | | Descriptor: | Mediator of RNA polymerase II transcription subunit 25, Non-structural protein 1 | | Authors: | Kalita, P, Borek, D.M, Amarasinghe, G.K, Leung, D.W. | | Deposit date: | 2024-06-23 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Molecular basis for human respiratory syncytial virus transcriptional regulator NS1 interactions with MED25.
Nat Commun, 16, 2025
|
|
6LDL
 
 | | Crystal structure of CYP116B46-N(20-445) from Tepidiphilus thermophilus in complex with HEME | | Descriptor: | BICINE, Cytochrome P450, GLYCEROL, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-11-21 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural insight into the electron transfer pathway of a self-sufficient P450 monooxygenase.
Nat Commun, 11, 2020
|
|
7M71
 
 | | SARS-CoV-2 Spike:5A6 Fab complex I focused refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 5A6 Fab heavy chain, Antibody 5A6 Fab light chain, ... | | Authors: | Asarnow, D, Cheng, Y. | | Deposit date: | 2021-03-26 | | Release date: | 2021-05-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural insight into SARS-CoV-2 neutralizing antibodies and modulation of syncytia.
Cell, 184, 2021
|
|
7M7B
 
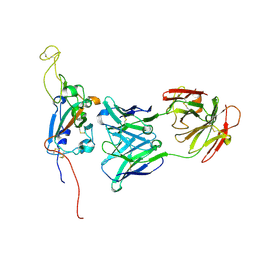 | | SARS-CoV-2 Spike:Fab 3D11 complex focused refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab 3D11 heavy chain, Antibody Fab 3D11 light chain, ... | | Authors: | Asarnow, D, Cheng, Y. | | Deposit date: | 2021-03-27 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural insight into SARS-CoV-2 neutralizing antibodies and modulation of syncytia.
Cell, 184, 2021
|
|
5KUC
 
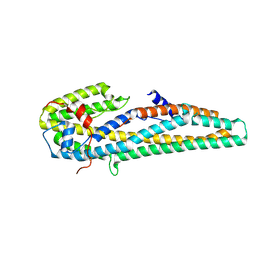 | | Crystal structure of trypsin activated Cry6Aa | | Descriptor: | Pesticidal crystal protein Cry6Aa | | Authors: | Kelker, M.S, Xu, X, Lee, M, Chan, M, Hung, S, Dementiev, K, Hey, T, Chikwana, V.M, Narva, K.E. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The pesticidal Cry6Aa toxin from Bacillus thuringiensis is structurally similar to HlyE-family alpha pore-forming toxins.
Bmc Biol., 14, 2016
|
|
5KUD
 
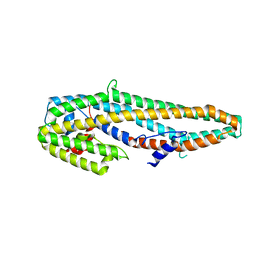 | | Crystal structure of full length Cry6Aa | | Descriptor: | Pesticidal crystal protein Cry6Aa | | Authors: | Kelker, M.S, Xu, X, Lee, M, Chan, M, Hung, S, Dementiev, K, Chikwana, V.M, Hey, T, Narva, K. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The pesticidal Cry6Aa toxin from Bacillus thuringiensis is structurally similar to HlyE-family alpha pore-forming toxins.
Bmc Biol., 14, 2016
|
|
4TQK
 
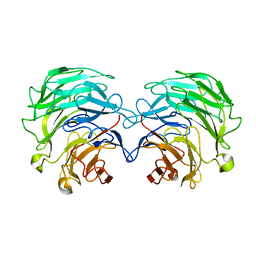 | | Structural basis of specific recognition of non-reducing terminal N-acetylglucosamine by an Agrocybe aegerita lection | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lectin 2 | | Authors: | Hu, Y.L, Ren, X.M, Li, D.F, Jiang, S, Lan, X.Q, Sun, H, Wang, D.C. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Specific Recognition of Non-Reducing Terminal N-Acetylglucosamine by an Agrocybe aegerita Lectin.
Plos One, 10, 2015
|
|
8VZD
 
 | | Crystal Structure of 2-Hydroxyacyl-CoA lyase/synthase TbHACS from Thermoflexaceae bacterium in the Complex with THDP, Formyl-CoA and ADP | | Descriptor: | 1,2-ETHANEDIOL, 2-Hydroxyacyl-CoA Lyase/Synthase TbHACS, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kim, Y, Maltseva, M, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
8VZB
 
 | | Crystal Structure of 2-Hydroxacyl-CoA Lyase/Synthase ApbHACS from Alphaproteobacteria bacterium in the Complex with THDP, D-Lactyl-CoA, and ADP | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2R)-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-2-oxidanyl-2H-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, ACETIC ACID, ... | | Authors: | Gade, P, Kim, Y, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
8VZJ
 
 | | Crystal Structure of 2-Hydroxyacyl-CoA Lyase/Synthase DhcHACS from Dehalococcoidia bacterium in the Complex with THDP and ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Kim, Y, Gade, P, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
8VZF
 
 | | Crystal Structure of 2-Hydroxyacyl-CoA Lyase/Synthase ApbHACS from Alphaproteobacteria bacterium in the Complex with THDP, Coenzyme A, and ADP | | Descriptor: | 1,2-ETHANEDIOL, 2-Hydroxyacyl-CoA Lyase/Synthase, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gade, P, Kim, Y, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
8VZH
 
 | | Crystal Structure of 2-Hydroxyacyl-CoA lyase/synthase TbHACS from Thermoflexaceae bacterium in the Complex with THDP and ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, M, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
8VZK
 
 | | Crystal Structure of 2-Hydroxyacyl-CoA Lyase/Synthase CfhHACS from Chloroflexi bacterium in the Complex with THDP and ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Gade, P, Mayfield, A, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
8VZI
 
 | | Crystal Structure of 2-Hydroxyacyl-CoA Lyase/Synthase CcHACS from Conidiobolus coronatus in the Complex with THDP and ADP | | Descriptor: | 1,2-ETHANEDIOL, 2-hydroxyacyl-CoA lyase-like protein 1, ACETATE ION, ... | | Authors: | Kim, Y, Gade, P, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
8VZC
 
 | | Crystal Structure of 2-Hydroxyacyl-CoA Lyase/Synthse ApbHACS from Alphaproteobacteria bacterium in the Complex with THDP, Formyl-CoA, and ADP | | Descriptor: | 1,2-ETHANEDIOL, 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-(hydroxymethyl)-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gade, P, Kim, Y, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
8VZA
 
 | | Crystal Structure of 2-Hydroxacyl-CoA Lyase/Synthase ApbHACS from Alphaproteobacteria bacterium in the Complex with THDP, L-Lactyl-CoA, and ADP | | Descriptor: | 2-Hydroxyacyl-CoA lyase/Synthase, 2-[(2R)-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-2-oxidanyl-2H-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, ACETYL GROUP, ... | | Authors: | Gade, P, Kim, Y, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
8VZE
 
 | | Crystal Structure of 2-Hydroxyacyl-CoA lyase/synthase TbHACS from Thermoflexaceae bacterium in the Complex with THDP, 2-Hydroxyisobutyryl-CoA and ADP | | Descriptor: | 1,2-ETHANEDIOL, 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-(hydroxymethyl)-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kim, Y, Maltseva, M, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
8GYM
 
 | | Cryo-EM structure of Tetrahymena thermophila respiratory mega-complex MC IV2+(I+III2+II)2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Wu, M.C, Hu, Y.Q, Han, F.Z, Zhou, L. | | Deposit date: | 2022-09-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structures of Tetrahymena thermophila respiratory megacomplexes on the tubular mitochondrial cristae.
Nat Commun, 14, 2023
|
|
4TQJ
 
 | | Structural basis of specific recognition of non-reducing terminal N-acetylglucosamine by an Agrocybe aegerita lection | | Descriptor: | Lectin 2 | | Authors: | Hu, Y.L, Ren, X.M, Li, D.F, Jiang, S, Lan, X.Q, Sun, H, Wang, D.C. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Specific Recognition of Non-Reducing Terminal N-Acetylglucosamine by an Agrocybe aegerita Lectin.
Plos One, 10, 2015
|
|
4TQM
 
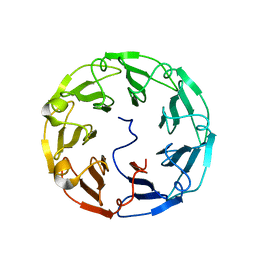 | | Structural basis of specific recognition of non-reducing terminal N-acetylglucosamine by an Agrocybe aegerita lection | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lectin 2 | | Authors: | Hu, Y.L, Ren, X.M, Li, D.F, Jiang, S, Lan, X.Q, Sun, H, Wang, D.C. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Specific Recognition of Non-Reducing Terminal N-Acetylglucosamine by an Agrocybe aegerita Lectin.
Plos One, 10, 2015
|
|
7KQB
 
 | | SARS-CoV-2 spike glycoprotein:Fab 5A6 complex I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 5A6 heavy chain, Fab 5A6 light chain, ... | | Authors: | Asarnow, D, Charles, C, Cheng, Y. | | Deposit date: | 2020-11-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structural insight into SARS-CoV-2 neutralizing antibodies and modulation of syncytia.
Cell, 184, 2021
|
|
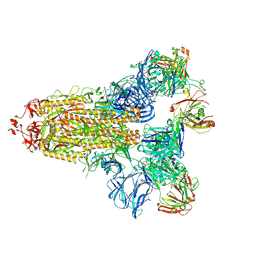
7KQE
 
 | |
