7SIU
 
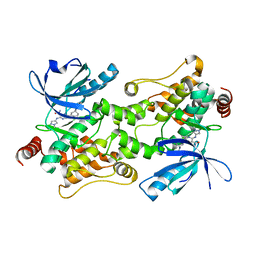 | | Crystal structure of HPK1 (MAP4K1) complex with inhibitor A-745 | | Descriptor: | 6-[(5R)-5-benzamidocyclohex-1-en-1-yl]-3-[(1-methyl-1H-pyrazol-4-yl)amino]pyrazine-2-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1, SULFATE ION | | Authors: | Longenecker, K.L, Korepanova, A, Qiu, W. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | The HPK1 Inhibitor A-745 Verifies the Potential of Modulating T Cell Kinase Signaling for Immunotherapy.
Acs Chem.Biol., 17, 2022
|
|
6VKV
 
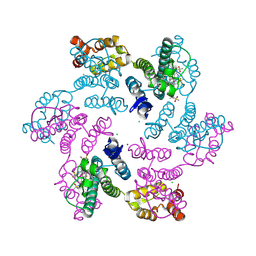 | |
8XKF
 
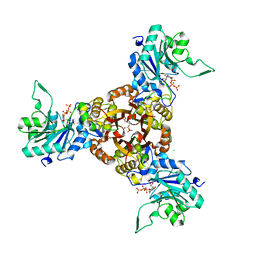 | | Crystal structure of Helicobacter pylori IspDF with substrate CTP | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional enzyme IspD/IspF, CHLORIDE ION, ... | | Authors: | Chen, X, Wu, D. | | Deposit date: | 2023-12-23 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two natural compounds as potential inhibitors against the Helicobacter pylori and Acinetobacter baumannii IspD enzymes.
Int J Antimicrob Agents, 63, 2024
|
|
5XS5
 
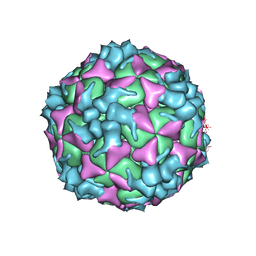 | | Structure of Coxsackievirus A6 (CVA6) virus procapsid particle | | Descriptor: | Genome polyprotein | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Cheng, T, Li, S.W. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
5XS7
 
 | | Structure of Coxsackievirus A6 (CVA6) virus A-particle in complex with the neutralizing antibody fragment 1D5 | | Descriptor: | Genome polyprotein, Heavy chain of Fab 1D5, Light chain of Fab 1D5 | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Li, S.W, Cheng, T. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
5XS4
 
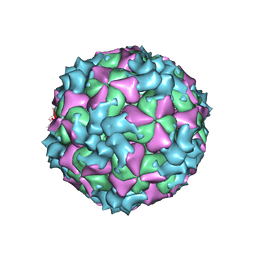 | | Structure of Coxsackievirus A6 (CVA6) virus A-particle | | Descriptor: | Genome polyprotein | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Li, S.W, Cheng, T. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
4YH3
 
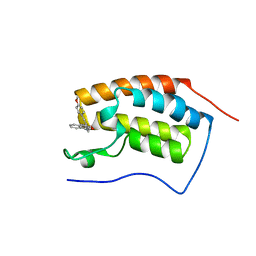 | | Crystal structure of human BRD4(1) in complex with 4-[(2E)-3-(4-methoxyphenyl)-2-phenylprop-2-enoyl]-3,4-dihydroquinoxalin-2(1H)-one (compound 19a) | | Descriptor: | 4-[(2E)-3-(4-methoxyphenyl)-2-phenylprop-2-enoyl]-3,4-dihydroquinoxalin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | White, A, Lakshminarasimhan, D, Suto, R.K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of a new chemical series of BRD4(1) inhibitors using protein-ligand docking and structure-guided design.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4YH4
 
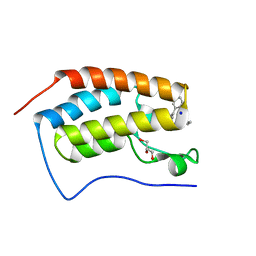 | | Crystal structure of human BRD4(1) in complex with 4-[(5-phenylpyridin-3-yl)carbonyl]-3,4-dihydroquinoxalin-2(1H)-one (compound 19d) | | Descriptor: | 4-[(5-phenylpyridin-3-yl)carbonyl]-3,4-dihydroquinoxalin-2(1H)-one, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of a new chemical series of BRD4(1) inhibitors using protein-ligand docking and structure-guided design.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3HWP
 
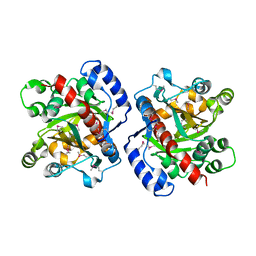 | | Crystal structure and computational analyses provide insights into the catalytic mechanism of 2, 4-diacetylphloroglucinol hydrolase PhlG from Pseudomonas fluorescens | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, PhlG, ... | | Authors: | He, Y.-X, Huang, L, Xue, Y, Fei, X, Teng, Y.-B, Zhou, C.-Z. | | Deposit date: | 2009-06-18 | | Release date: | 2009-12-15 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Computational Analyses Provide Insights into the Catalytic Mechanism of 2,4-Diacetylphloroglucinol Hydrolase PhlG from Pseudomonas fluorescens.
J.Biol.Chem., 285, 2010
|
|
4AME
 
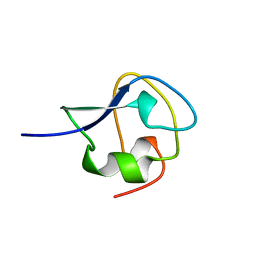 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T18A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
3M6I
 
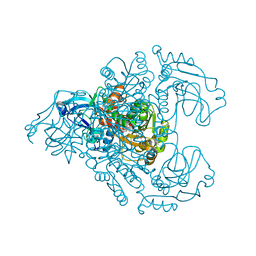 | | L-arabinitol 4-dehydrogenase | | Descriptor: | L-arabinitol 4-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2010-03-15 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and engineering of L-arabinitol 4-dehydrogenase from Neurospora crassa
J.Mol.Biol., 402, 2010
|
|
5IVX
 
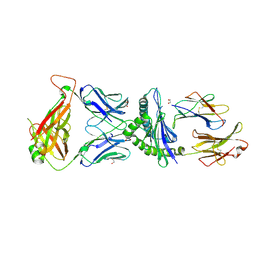 | | Crystal Structure of B4.2.3 T-Cell Receptor and H2-Dd P18-I10 Complex | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Natarajan, K, Jiang, J, Margulies, D. | | Deposit date: | 2016-03-21 | | Release date: | 2017-03-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An allosteric site in the T-cell receptor C beta domain plays a critical signalling role.
Nat Commun, 8, 2017
|
|
5IW1
 
 | |
3OHI
 
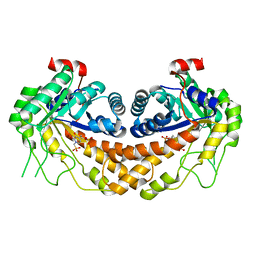 | | Structure of Giardia fructose-1,6-biphosphate aldolase in complex with 3-hydroxy-2-pyridone | | Descriptor: | ({3-hydroxy-2-oxo-4-[2-(phosphonooxy)ethyl]pyridin-1(2H)-yl}methyl)phosphonic acid, Putative fructose-1,6-bisphosphate aldolase, ZINC ION | | Authors: | Herzberg, O, Galkin, A. | | Deposit date: | 2010-08-17 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational design, synthesis and evaluation of first generation inhibitors of the Giardia lamblia fructose-1,6-biphosphate aldolase.
J.Inorg.Biochem., 105, 2010
|
|
7JI3
 
 | |
3EWH
 
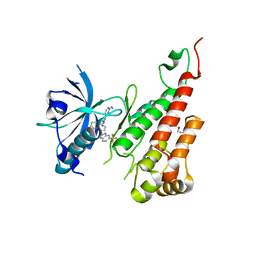 | | Crystal structure of the VEGFR2 kinase domain in complex with a pyridyl-pyrimidine benzimidazole inhibitor | | Descriptor: | 1,2-ETHANEDIOL, N-[4-({3-[2-(methylamino)pyrimidin-4-yl]pyridin-2-yl}oxy)naphthalen-1-yl]-6-(trifluoromethyl)-1H-benzimidazol-2-amine, vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2008-10-15 | | Release date: | 2009-08-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pyridyl-pyrimidine benzimidazole derivatives as potent, selective, and orally bioavailable inhibitors of Tie-2 kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1MSJ
 
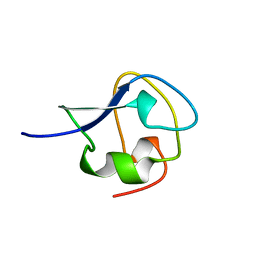 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T15V | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
7K9K
 
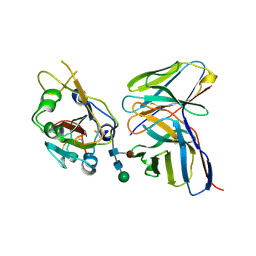 | |
7K9H
 
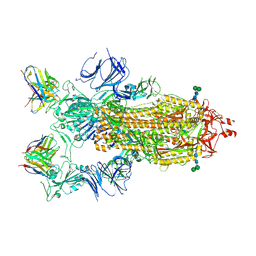 | | SARS-CoV-2 Spike in complex with neutralizing Fab 2B04 (one up, two down conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B04 heavy chain, ... | | Authors: | Errico, J.M, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mechanism of SARS-CoV-2 neutralization by two murine antibodies targeting the RBD.
Cell Rep, 37, 2021
|
|
7K9I
 
 | |
7K9J
 
 | |
6AME
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 M21A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1B7J
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 V20A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
7JUO
 
 | | CBP bromodomain complexed with YF2-23 | | Descriptor: | CREB-binding protein, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-(1-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-3-yl)-1H-indol-4-yl}ethanesulfonamide | | Authors: | Ratia, K.M, Xiong, R, Principe, D, Li, Y, Huang, F, Rana, A, Thatcher, G. | | Deposit date: | 2020-08-20 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | XP-524 is a dual-BET/EP300 inhibitor that represses oncogenic KRAS and potentiates immune checkpoint inhibition in pancreatic cancer.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8GU0
 
 | | Crystal structure of a fungal halogenase RadH | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Non-heme halogenase radH, ... | | Authors: | Jiang, S.M, Brown, C.J. | | Deposit date: | 2022-09-09 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Further Characterization of Fungal Halogenase RadH and Its Homologs.
Biomolecules, 13, 2023
|
|
