7Y7M
 
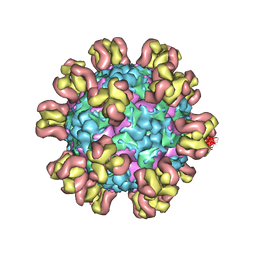 | |
7XY9
 
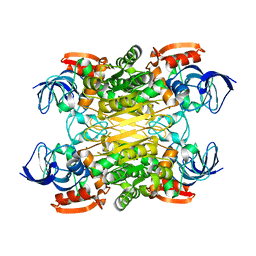 | | Cryo-EM structure of secondary alcohol dehydrogenases TbSADH after carrier-free immobilization based on weak intermolecular interactions | | Descriptor: | MAGNESIUM ION, NADP-dependent isopropanol dehydrogenase, ZINC ION | | Authors: | Chen, Q, Li, X, Yang, F, Qu, G, Sun, Z.T, Wang, Y.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Active and stable alcohol dehydrogenase-assembled hydrogels via synergistic bridging of triazoles and metal ions.
Nat Commun, 14, 2023
|
|
7YHH
 
 | |
7YHG
 
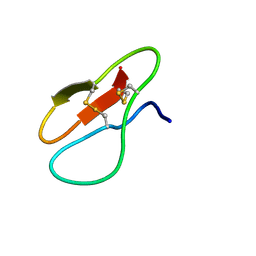 | |
7YHF
 
 | |
7YHI
 
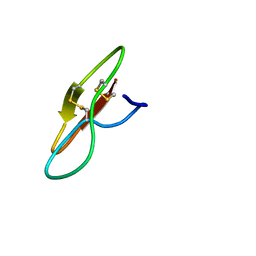 | |
7YRH
 
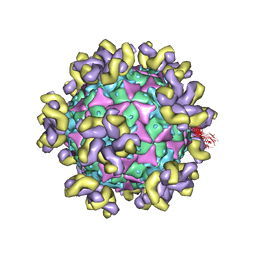 | |
7YRF
 
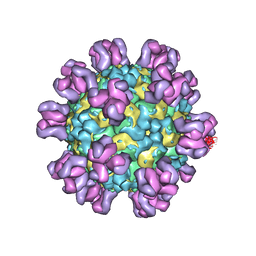 | |
7YV7
 
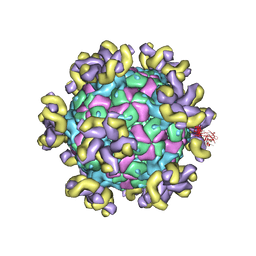 | |
7YV2
 
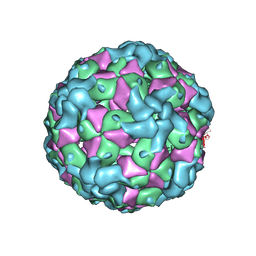 | |
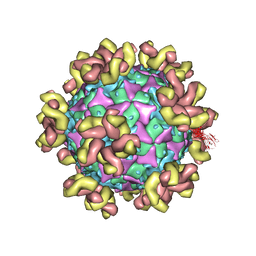
7YMS
 
 | |
7YOW
 
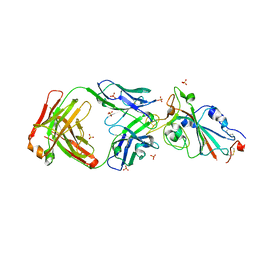 | |
7XGW
 
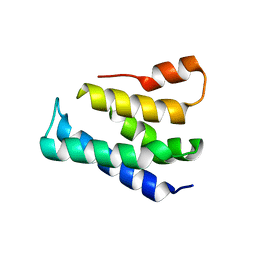 | |
7WVR
 
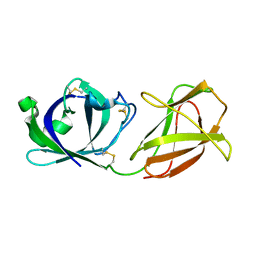 | |
7X1R
 
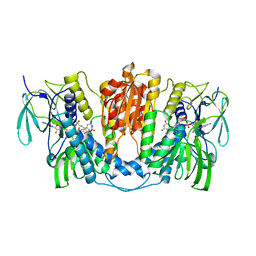 | | Cryo-EM structure of human thioredoxin reductase bound by Au | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GOLD ION, Thioredoxin reductase 1, ... | | Authors: | He, Z.S, Cao, P, Cao, S.H, He, B, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2022-02-24 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Au4 cluster inhibits human thioredoxin reductase activity via specifically binding of Au to Cys189
Nano Today, 47, 2022
|
|
7XAC
 
 | |
7XBL
 
 | |
5TO3
 
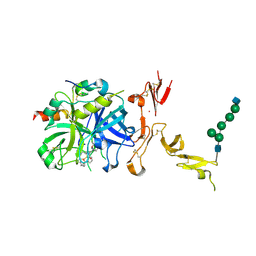 | | Crystal structure of thrombin mutant W215A/E217A fused to EGF456 of thrombomodulin via a 31-residue linker and bound to PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose-(1-4)-[beta-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Barranco-Medina, S, Murphy, M, Pelc, L, Chen, Z, Di Cera, E, Pozzi, N. | | Deposit date: | 2016-10-16 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Rational Design of Protein C Activators.
Sci Rep, 7, 2017
|
|
3WAJ
 
 | |
3WAK
 
 | |
7BYL
 
 | | Cryo-EM structure of human KCNQ4 | | Descriptor: | Calmodulin-3, Green fluorescent protein,Potassium voltage-gated channel subfamily KQT member 4, POTASSIUM ION, ... | | Authors: | Shen, H, Li, T, Yue, Z. | | Deposit date: | 2020-04-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural Basis for the Modulation of Human KCNQ4 by Small-Molecule Drugs.
Mol.Cell, 81, 2021
|
|
7BYN
 
 | | Cryo-EM structure of human KCNQ4 with linopirdine | | Descriptor: | 1-phenyl-3,3-bis(pyridin-4-ylmethyl)indol-2-one, Calmodulin-3, Green fluorescent protein,Potassium voltage-gated channel subfamily KQT member 4, ... | | Authors: | Shen, H, Li, T, Yue, Z. | | Deposit date: | 2020-04-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Basis for the Modulation of Human KCNQ4 by Small-Molecule Drugs.
Mol.Cell, 81, 2021
|
|
7CCE
 
 | |
7CC9
 
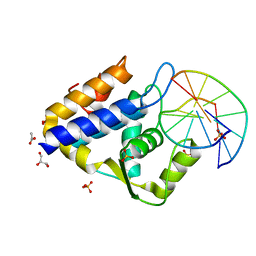 | | Sulfur binding domain of SprMcrA complexed with phosphorothioated DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*GP*GP*CP*GP*GS*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*CP*CP*GP*CP*C)-3'), ... | | Authors: | Yu, H, Zhao, G, Gan, J, Liu, G, Wu, G, He, X. | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.063 Å) | | Cite: | DNA backbone interactions impact the sequence specificity of DNA sulfur-binding domains: revelations from structural analyses.
Nucleic Acids Res., 48, 2020
|
|
7CCJ
 
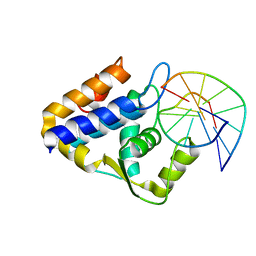 | | Sulfur binding domain of SprMcrA complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(*GP*GP*AP*TP*CP*AP*TP*C)-3'), HNHc domain-containing protein | | Authors: | Yu, H, Zhao, G, Gan, J, Liu, G, Wu, G, He, X. | | Deposit date: | 2020-06-17 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DNA backbone interactions impact the sequence specificity of DNA sulfur-binding domains: revelations from structural analyses.
Nucleic Acids Res., 48, 2020
|
|
