6SFI
 
 | | Crystal structure of p38 alpha in complex with compound 75 (MCP33) | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]-2-[[[1-(2-methylphenyl)pyrazol-4-yl]carbonylamino]methyl]-1,3-thiazole-5-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
4P7A
 
 | | Crystal Structure of human MLH1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein Mlh1, MAGNESIUM ION, ... | | Authors: | Tempel, W, Lam, R, Zeng, H, Walker, J.R, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the human MLH1 N-terminus: implications for predisposition to Lynch syndrome.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6SFJ
 
 | | Crystal structure of p38 alpha in complex with compound 77 (MCP41) | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[5-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]-2-methyl-phenyl]-1-(2-methylphenyl)pyrazole-4-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6SJM
 
 | | Crystal structure of the Retinoic Acid Receptor alpha in complex with compound 24 (JP175) | | Descriptor: | 2-[4-[3,5-bis(trifluoromethyl)phenyl]phenyl]ethanoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Chaikuad, A, Pollinger, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-13 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | A Novel Biphenyl-based Chemotype of Retinoid X Receptor Ligands Enables Subtype and Heterodimer Preferences.
Acs Med.Chem.Lett., 10, 2019
|
|
4QSQ
 
 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) with bound DMSO | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
6SE4
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with (+)-JD1, an Organometallic BET Bromodomain Inhibitor | | Descriptor: | (+)-JD1, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Krojer, T, Hassell-Hart, S, Picaud, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Spencer, J, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD4 in complex with (+)-JD1, an Organometallic BET Bromodomain Inhibitor
To Be Published
|
|
4QQK
 
 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) with GMS | | Descriptor: | (5S)-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}-N~6~-carbamimidoyl-L-lysine, GLYCEROL, Protein arginine N-methyltransferase 6, ... | | Authors: | Dong, A, Zeng, H, He, H, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
3EBB
 
 | | PLAP/P97 complex | | Descriptor: | MAGNESIUM ION, PHOSPHOLIPASE A2-ACTIVATING PROTEIN, TRANSITIONAL ENDOPLASMIC RETICULUM ATPASE (TER ATP | | Authors: | Walker, J.R, Qiu, L, Akutsu, M, Slessarev, Y, Amaya, M.F, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-27 | | Release date: | 2009-02-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the PLAA/Ufd3-p97/Cdc48 complex.
J.Biol.Chem., 285, 2010
|
|
3EP0
 
 | | Methyltransferase domain of human PR domain-containing protein 12 | | Descriptor: | PR domain zinc finger protein 12 | | Authors: | Amaya, M.F, Zeng, H, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Min, J, Plotnikov, A.N, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of methyltransferase domain of human PR
Domain-containing protein 12.
To be Published
|
|
3EO3
 
 | | Crystal structure of the N-acetylmannosamine kinase domain of human GNE protein | | Descriptor: | Bifunctional UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Nedyalkova, L, Tong, Y, Rabeh, W.M, Hong, B, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-26 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of the N-acetylmannosamine kinase domain of GNE.
Plos One, 4, 2009
|
|
6R7Y
 
 | | CryoEM structure of calcium-bound human TMEM16K / Anoctamin 10 in detergent (low Ca2+, closed form) | | Descriptor: | Anoctamin-10, CALCIUM ION | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
6R65
 
 | | Crystal Structure of human TMEM16K / Anoctamin 10 (Form 2) | | Descriptor: | Anoctamin-10, CALCIUM ION | | Authors: | Bushell, S.R, Pike, A.C.W, Chu, A, Tessitore, A, Rotty, B, Mukhopadhyay, S, Kupinska, K, Shrestha, L, Borkowska, O, Chalk, R, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-26 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
3EMW
 
 | | Crystal Structure of human splA/ryanodine receptor domain and SOCS box containing 2 (SPSB2) in complex with a 20-residue VASA peptide | | Descriptor: | 1,2-ETHANEDIOL, Peptide (VASA), SPRY domain-containing SOCS box protein 2 | | Authors: | Filippakopoulos, P, Sharpe, T, Keates, T, Murray, J.W, Savitsky, P, Roos, A.K, Pike, A.C.W, Von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for Par-4 recognition by the SPRY domain- and SOCS box-containing proteins SPSB1, SPSB2, and SPSB4.
J.Mol.Biol., 401, 2010
|
|
6T1M
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-cyano-~{N}-[2-(piperidin-1-ylmethyl)-1~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
4QSW
 
 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with 5-methyl uridine | | Descriptor: | 1,2-ETHANEDIOL, 5-methyluridine, ATPase family AAA domain-containing protein 2, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
6T1I
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with piperazine-urea derivative 1 | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-ethanoylphenyl)-~{N}-[(6-methoxypyridin-3-yl)methyl]piperazine-1-carboxamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6T1O
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 6 | | Descriptor: | 1,2-ETHANEDIOL, 4-iodanyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6SFK
 
 | | Crystal structure of p38 alpha in complex with compound 81 (MCP42) | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 14, ~{N}-[5-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]-2-methyl-phenyl]-1-phenyl-5-(trifluoromethyl)pyrazole-4-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6R7O
 
 | | Crystal structure of the central region of human cohesin subunit STAG1 | | Descriptor: | Cohesin subunit SA-1 | | Authors: | Newman, J.A, katis, V.L, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-03-29 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | STAG1 vulnerabilities for exploiting cohesin synthetic lethality in STAG2-deficient cancers.
Life Sci Alliance, 3, 2020
|
|
3FEH
 
 | | Crystal structure of full length centaurin alpha-1 | | Descriptor: | Centaurin-alpha-1, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Shen, L, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-29 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phosphorylation-independent dual-site binding of the FHA domain of KIF13 mediates phosphoinositide transport via centaurin {alpha}1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6R7X
 
 | | CryoEM structure of calcium-bound human TMEM16K / Anoctamin 10 in detergent (2mM Ca2+, closed form) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-10, CALCIUM ION, ... | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Baronina, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
3FE4
 
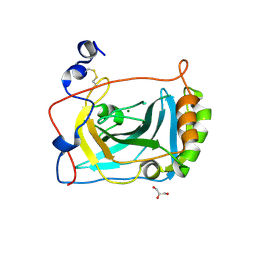 | | Crystal Structure of Human Carbonic Anhydrase vi | | Descriptor: | Carbonic anhydrase 6, GLYCEROL, MAGNESIUM ION | | Authors: | Pilka, E.S, Kochan, G, Krysztofinska, E, Muniz, J, Yue, W.W, Roos, A.K, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-27 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the secretory isozyme of mammalian carbonic anhydrases CA VI: implications for biological assembly and inhibitor development
Biochem.Biophys.Res.Commun., 419, 2012
|
|
3FEO
 
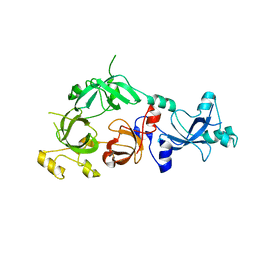 | | The crystal structure of MBTD1 | | Descriptor: | MBT domain-containing protein 1 | | Authors: | Amaya, M.F, Eryilmaz, J, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-30 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies of a four-MBT repeat protein MBTD1.
Plos One, 4, 2009
|
|
3FLV
 
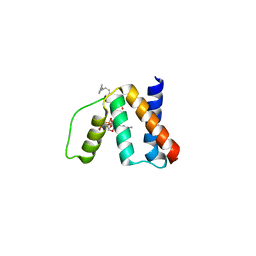 | | The crystal structure of human acyl-CoenzymeA binding domain containing 5 | | Descriptor: | Acyl-CoA-binding domain-containing protein 5, COENZYME A, STEARIC ACID, ... | | Authors: | Ugochukwu, E, Roos, A, Yue, W.W, Shafqat, N, Salah, E, Savitsky, P, Muniz, J.R.C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-19 | | Release date: | 2009-02-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of human acyl-Coenzyme A binding domain containing 5
To be Published
|
|
4QSU
 
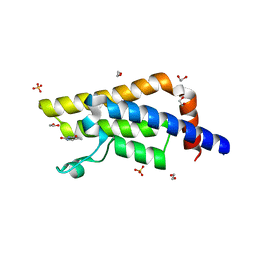 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with thymine | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, SULFATE ION, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
