6E83
 
 | |
1FTA
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE, 1-PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH THE ALLOSTERIC INHIBITOR AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Lipscomb, W.N. | | Deposit date: | 1993-09-27 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The allosteric site of human liver fructose-1,6-bisphosphatase. Analysis of six AMP site mutants based on the crystal structure.
J.Biol.Chem., 269, 1994
|
|
4XV3
 
 | | B-Raf Kinase V600E oncogenic mutant in complex with PLX7922 | | Descriptor: | N'-{3-[5-(2-aminopyrimidin-4-yl)-2-tert-butyl-1,3-thiazol-4-yl]-2-fluorophenyl}-N-ethyl-N-methylsulfuric diamide, Serine/threonine-protein kinase B-raf | | Authors: | Zhang, Y, Zhang, C. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RAF inhibitors that evade paradoxical MAPK pathway activation.
Nature, 526, 2015
|
|
4XV9
 
 | | B-Raf Kinase domain in complex with PLX5568 | | Descriptor: | N-{3-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)carbonyl]-2,4-difluorophenyl}-4-(trifluoromethyl)benzenesulfonamide, SULFATE ION, Serine/threonine-protein kinase B-raf | | Authors: | zhang, Y, zhang, c, wang, w. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RAF inhibitors that evade paradoxical MAPK pathway activation.
Nature, 526, 2015
|
|
4XV1
 
 | | B-Raf Kinase V600E oncogenic mutant in complex with PLX7904 | | Descriptor: | N'-(3-{[5-(2-cyclopropylpyrimidin-5-yl)-1H-pyrrolo[2,3-b]pyridin-3-yl]carbonyl}-2,4-difluorophenyl)-N-ethyl-N-methylsulfuric diamide, Serine/threonine-protein kinase B-raf | | Authors: | Zhang, Y, Zhang, C. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | RAF inhibitors that evade paradoxical MAPK pathway activation.
Nature, 526, 2015
|
|
4XV2
 
 | |
2VI5
 
 | | LUMAZINE SYNTHASE FROM MYCOBACTERIUM TUBERCULOSIS BOUND TO N-6-(ribitylamino)pyrimidine-2,4(1H,3H)-dione-5-yl-propionamide | | Descriptor: | 1-deoxy-1-{[(5S)-2,6-dioxo-5-(propanoylamino)-1,2,5,6-tetrahydropyrimidin-4-yl]amino}-D-ribitol, 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, PHOSPHATE ION, ... | | Authors: | Morgunova, E, Zhang, Y, Jin, G, Illarionov, B, Bacher, A, Fischer, M, Cushman, M, Ladenstein, R. | | Deposit date: | 2007-11-27 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A New Series of N-[2,4-Dioxo-6-D-Ribitylamino-1,2, 3,4-Tetrahydropyrimidin-5-Yl]Oxalamic Acid Derivatives as Inhibitors of Lumazine Syntase and Riboflavin Synthase: Design, Synthesis, Biochemical Evaluation, Crystallography and Mechanistic Implications.
J.Org.Chem., 73, 2008
|
|
3OG7
 
 | | B-Raf Kinase V600E oncogenic mutant in complex with PLX4032 | | Descriptor: | AKAP9-BRAF fusion protein, N-(3-{[5-(4-chlorophenyl)-1H-pyrrolo[2,3-b]pyridin-3-yl]carbonyl}-2,4-difluorophenyl)propane-1-sulfonamide | | Authors: | Zhang, Y, Zhang, K.Y, Zhang, C. | | Deposit date: | 2010-08-16 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma.
Nature, 467, 2010
|
|
4XVC
 
 | |
4R7H
 
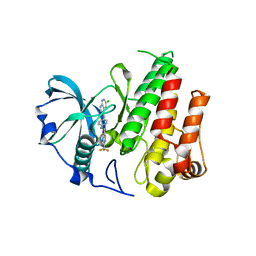 | | Crystal structure of FMS KINASE domain with a small molecular inhibitor, PLX3397 | | Descriptor: | 5-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)methyl]-N-{[6-(trifluoromethyl)pyridin-3-yl]methyl}pyridin-2-amine, Macrophage colony-stimulating factor 1 receptor | | Authors: | Zhang, Y, Zhang, K, Zhang, C. | | Deposit date: | 2014-08-27 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8001 Å) | | Cite: | Structure-Guided Blockade of CSF1R Kinase in Tenosynovial Giant-Cell Tumor.
N Engl J Med, 373, 2015
|
|
4R7I
 
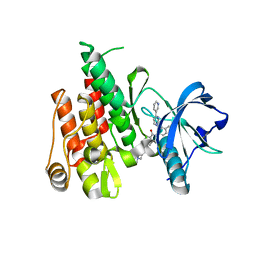 | |
4RT7
 
 | | Crystal Structure of FLT3 with a small molecule inhibitor | | Descriptor: | 1-(5-tert-butyl-1,2-oxazol-3-yl)-3-(4-{7-[2-(morpholin-4-yl)ethoxy]imidazo[2,1-b][1,3]benzothiazol-2-yl}phenyl)urea, Receptor-type tyrosine-protein kinase FLT3 | | Authors: | Zhang, Y, Zhang, C. | | Deposit date: | 2014-11-13 | | Release date: | 2015-04-22 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Characterizing and Overriding the Structural Mechanism of the Quizartinib-Resistant FLT3 "Gatekeeper" F691L Mutation with PLX3397.
Cancer Discov, 5, 2015
|
|
6O5W
 
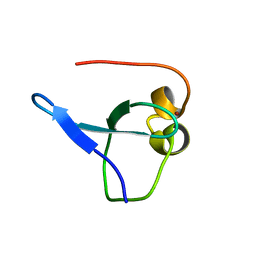 | | Crystal structure of MORC3 CW domain fused with viral influenza A NS1 peptide | | Descriptor: | NS1-linked peptide,MORC family CW-type zinc finger protein 3, ZINC ION | | Authors: | Ahn, J, Zhang, Y, Vann, K.R, Kutateleadze, T. | | Deposit date: | 2019-03-04 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.412 Å) | | Cite: | MORC3 Is a Target of the Influenza A Viral Protein NS1.
Structure, 27, 2019
|
|
6P79
 
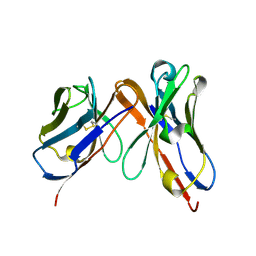 | | Engineered single chain antibody C9+C14 ScFv | | Descriptor: | Engineered antibody heavy chain, Engineered antibody light chain | | Authors: | Zhang, Y, Li, W, Marshall, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.583 Å) | | Cite: | Computer-based Engineering of Thermostabilized Antibody Fragments.
Aiche J, 66, 2020
|
|
1R0V
 
 | |
1R11
 
 | |
8ZMR
 
 | | Vesamicol-bound VAChT | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Vesicular acetylcholine transporter,DARPinoff7, vesamicol | | Authors: | Zhang, Z, Zhang, Y, Dai, F, Zhang, Y.X, Lee, C.-H. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into VAChT neurotransmitter recognition and inhibition.
Cell Res., 2024
|
|
8ZMS
 
 | | Acetylcholine-bound VAChT | | Descriptor: | ACETYLCHOLINE, Maltose/maltodextrin-binding periplasmic protein,Vesicular acetylcholine transporter,DARPinoff7 | | Authors: | Zhang, Z, Zhang, Y, Dai, F, Zhang, Y.X, Lee, C.-H. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into VAChT neurotransmitter recognition and inhibition.
Cell Res., 2024
|
|
5E17
 
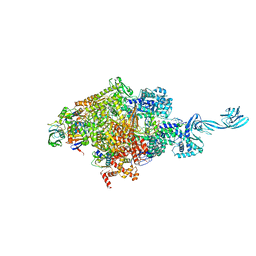 | |
4HW7
 
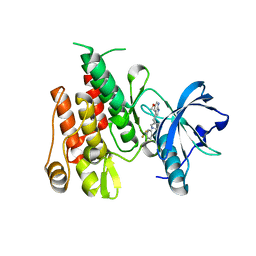 | |
4HVS
 
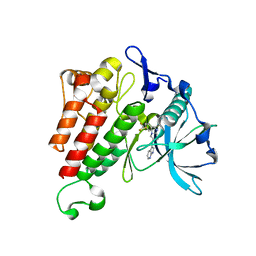 | |
4LA2
 
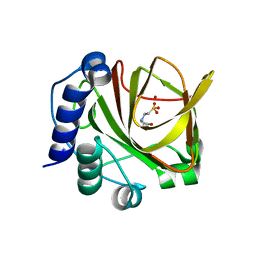 | | Crystal structure of dimethylsulphoniopropionate (DMSP) lyase DddQ | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Dimethylsulphoniopropionate (DMSP) lyase DddQ, ZINC ION | | Authors: | Zhang, Y, Li, C. | | Deposit date: | 2013-06-19 | | Release date: | 2014-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insight into bacterial cleavage of oceanic dimethylsulfoniopropionate into dimethyl sulfide
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5COI
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1-({[(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)sulfonyl]amino}methyl)cyclopentanecarboxylic acid, Bromodomain-containing protein 4 | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CPE
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-cycloheptyl-1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indole-6-sulfonamide | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-21 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CP5
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, 1-ethyl-N-(4-fluorophenyl)-2-oxo-1,2-dihydrobenzo[cd]indole-6-sulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-21 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
