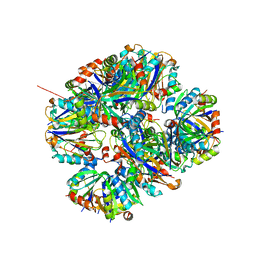
1F1O
 
 | | STRUCTURAL STUDIES OF ADENYLOSUCCINATE LYASES | | Descriptor: | ADENYLOSUCCINATE LYASE | | Authors: | Toth, E.A, Yeates, T. | | Deposit date: | 2000-05-19 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The crystal structure of adenylosuccinate lyase from Pyrobaculum aerophilum reveals an intracellular protein with three disulfide bonds.
J.Mol.Biol., 301, 2000
|
|
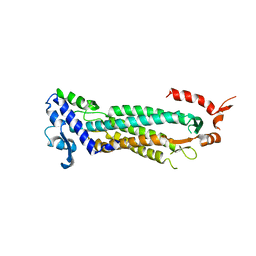
4ZK7
 
 | | Crystal structure of rescued two-component self-assembling tetrahedral cage T33-31 | | Descriptor: | Chorismate mutase, Divalent-cation tolerance protein CutA | | Authors: | Liu, Y, Cascio, D, Sawaya, M.R, Bale, J, Collazo, M.J, Park, R, King, N, Baker, D, Yeates, T. | | Deposit date: | 2015-04-30 | | Release date: | 2015-07-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a designed tetrahedral protein assembly variant engineered to have improved soluble expression.
Protein Sci., 24, 2015
|
|
5HS0
 
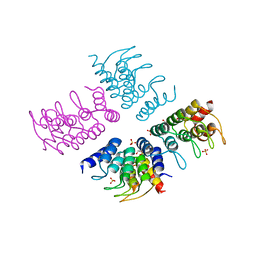 | | Computationally Designed Cyclic Tetramer ank1C4_7 | | Descriptor: | Ankyrin domain protein ank1C4_7, GLYCEROL, SULFATE ION | | Authors: | McNamara, D.E, Cascio, D, Fallas, J.A, Baker, D, Yeates, T.O. | | Deposit date: | 2016-01-24 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
5HRZ
 
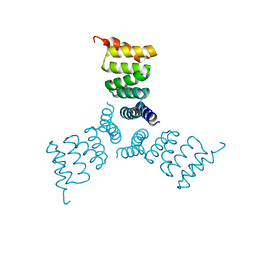 | | Computationally Designed Trimer 1na0C3_3 | | Descriptor: | TPR domain protein 1na0C3_3 | | Authors: | Cascio, D, McNamara, D.E, Fallas, J.A, Baker, D, Yeates, T.O. | | Deposit date: | 2016-01-24 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
5HRY
 
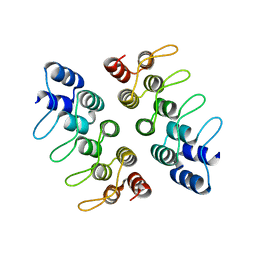 | | Computationally Designed Cyclic Dimer ank3C2_1 | | Descriptor: | ank3C2_1 | | Authors: | Cascio, D, McNamara, D.E, Fallas, J.A, Baker, D, Yeates, T.O. | | Deposit date: | 2016-01-24 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
4U9P
 
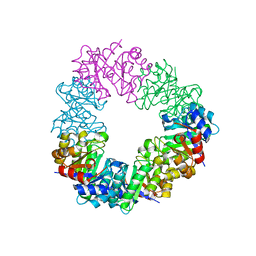 | | Structure of the methanofuran/methanopterin biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii | | Descriptor: | GLYCEROL, UPF0264 protein MJ1099 | | Authors: | Bobik, T.A, Morales, E, Shin, A, Cascio, D, Sawaya, M.R, Arbing, M, Rasche, M.E. | | Deposit date: | 2014-08-06 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the methanofuran/methanopterin-biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
2FGY
 
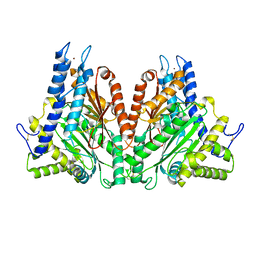 | |
2GM8
 
 | | TenA Homolog/Thi-4 Thiaminase complexed with product 4-amino-5-hydroxymethyl-2-methylpyrimidine | | Descriptor: | 1,2-ETHANEDIOL, 4-AMINO-5-HYDROXYMETHYL-2-METHYLPYRIMIDINE, tenA homolog/Thi-4 Thiaminase | | Authors: | Sawaya, M.R, Chan, S, Han, G.W, Perry, L.J, Pashkov, I. | | Deposit date: | 2006-04-06 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a TenA Homolog/Thi-4 Thiaminase from Pyrobaculum Aerophilum
To be Published
|
|
2GM7
 
 | | TenA Homolog/Thi-4 Thiaminase from Pyrobaculum Aerophilum | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Sawaya, M.R, Chan, S, Han, G.W, Perry, L.J. | | Deposit date: | 2006-04-06 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Ten A Homolog/Thi-4 Thiaminase from Pyrobaculum Aerophilum
To be Published
|
|
3NL9
 
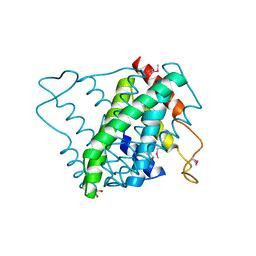 | |
1OEZ
 
 | | Zn His46Arg mutant of Human Cu, Zn Superoxide Dismutase | | Descriptor: | SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ZINC ION | | Authors: | Strange, R.W, Antonyuk, S, Hough, M.A, Doucette, P, Rodriguez, J, Elam, J.S, Hart, P.J, Hayward, L.J, Valentine, J.S, Hasnain, S.S. | | Deposit date: | 2003-04-02 | | Release date: | 2003-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Amyloid-Like Filaments and Water-Filled Nanotubes Formed by Sod1 Mutant Proteins Linked to Familial Als
Nat.Struct.Biol., 10, 2003
|
|
1MQ7
 
 | | CRYSTAL STRUCTURE OF DUTPASE FROM MYCOBACTERIUM TUBERCULOSIS (RV2697C) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Sawaya, M.R, Chan, S, Segelke, B.W, Lekin, T, Heike, K, Cho, U.S, Naranjo, C, Perry, L.J, Yeates, T.O, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-09-13 | | Release date: | 2002-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
1EAH
 
 | | PV2L COMPLEXED WITH ANTIVIRAL AGENT SCH48973 | | Descriptor: | 1[2-CHLORO-4-METHOXY-PHENYL-OXYMETHYL]-4-[2,6-DICHLORO-PHENYL-OXYMETHYL]-BENZENE, MYRISTIC ACID, POLIOVIRUS TYPE 2 COAT PROTEINS VP1 TO VP4 | | Authors: | Lentz, K, Arnold, E. | | Deposit date: | 1997-07-22 | | Release date: | 1998-09-16 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of poliovirus type 2 Lansing complexed with antiviral agent SCH48973: comparison of the structural and biological properties of three poliovirus serotypes.
Structure, 5, 1997
|
|
1NJ9
 
 | | Cocaine hydrolytic antibody 15A10 | | Descriptor: | SODIUM ION, immunoglobulin heavy chain, immunoglobulin variable chain | | Authors: | Larsen, N.A, de Prada, P, Deng, S.X, Zhu, X, Landry, D.W, Wilson, I.A. | | Deposit date: | 2002-12-30 | | Release date: | 2004-02-17 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystallographic and biochemical analysis of cocaine-degrading antibody 15A10.
Biochemistry, 43, 2004
|
|
3TUJ
 
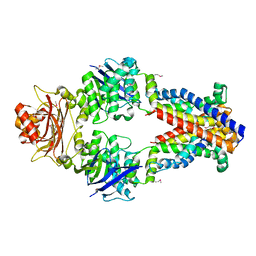 | |
3TUZ
 
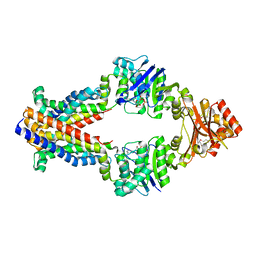 | | Inward facing conformations of the MetNI methionine ABC transporter: CY5 SeMet soak crystal form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-methionine transport system permease protein metI, Methionine import ATP-binding protein MetN, ... | | Authors: | Johnson, E, Nguyen, P, Rees, D.C. | | Deposit date: | 2011-09-19 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Inward facing conformations of the MetNI methionine ABC transporter: Implications for the mechanism of transinhibition.
Protein Sci., 21, 2012
|
|
3MG2
 
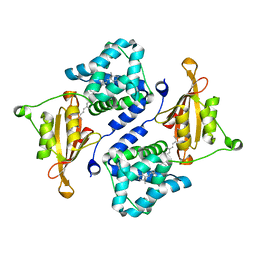 | | Crystal structure of the orange carotenoid protein Y44S mutant from cyanobacteria synechocystis sp. PCC 6803 | | Descriptor: | Orange carotenoid protein, beta,beta-caroten-4-one | | Authors: | Wilson, A, Kinney, J, Zwart, P.H, Punginelli, C, D'Haen, S, Perreau, F, Klein, M.G, Kirilovsky, D, Kerfeld, C.A. | | Deposit date: | 2010-04-05 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Structural determinants underlying photoprotection in the photoactive orange carotenoid protein of cyanobacteria.
J.Biol.Chem., 285, 2010
|
|
3MG1
 
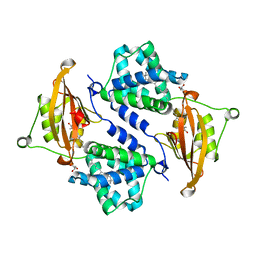 | | Crystal structure of the orange carotenoid protein from cyanobacteria Synechocystis sp. PCC 6803 | | Descriptor: | GLYCEROL, Orange carotenoid protein, beta,beta-caroten-4-one | | Authors: | Wilson, A, Kinney, J, Zwart, P.H, Punginelli, C, D'Haen, S, Perreau, F, Klein, M.G, Kirilovsky, D, Kerfeld, C.A. | | Deposit date: | 2010-04-05 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Structural determinants underlying photoprotection in the photoactive orange carotenoid protein of cyanobacteria.
J.Biol.Chem., 285, 2010
|
|
3MG3
 
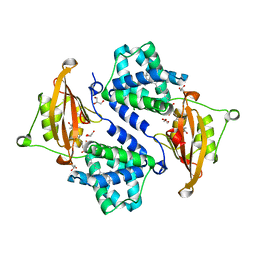 | | Crystal structure of the orange carotenoid protein R155L mutant from cyanobacteria synechocystis sp. PCC 6803 | | Descriptor: | GLYCEROL, Orange carotenoid-binding protein, beta,beta-caroten-4-one | | Authors: | Wilson, A, Kinney, J, Zwart, P.H, Punginelli, C, D'Haen, S, Perreau, F, Klein, M.G, Kirilovsky, D, Kerfeld, C.A. | | Deposit date: | 2010-04-05 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural determinants underlying photoprotection in the photoactive orange carotenoid protein of cyanobacteria.
J.Biol.Chem., 285, 2010
|
|
3TUI
 
 | |
2PLV
 
 | |
1SLH
 
 | | Mycobacterium tuberculosis dUTPase complexed with magnesium and dUDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.S, Kim, M.-Y, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-05 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
1SNF
 
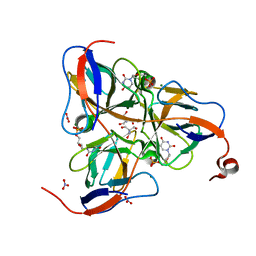 | | MYCOBACTERIUM TUBERCULOSIS DUTPASE COMPLEXED WITH MAGNESIUM AND DEOXYURIDINE 5'-MONOPHOSPHATE | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.S, Kim, M.-Y, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-10 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
1SIX
 
 | | Mycobacterium tuberculosis dUTPase complexed with magnesium and alpha,beta-imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.-S, Kim, M, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
1SM8
 
 | | M. tuberculosis dUTPase complexed with chromium and dUTP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHROMIUM ION, DEOXYURIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.S, Kim, M.-Y, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-08 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
