2ROO
 
 | |
1IDZ
 
 | |
1IDY
 
 | |
1WZE
 
 | |
1WZI
 
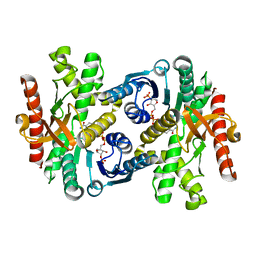 | |
1Y7T
 
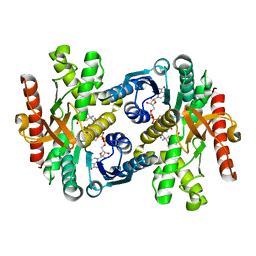 | | Crystal structure of NAD(H)-depenent malate dehydrogenase complexed with NADPH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Malate dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tomita, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2004-12-10 | | Release date: | 2005-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of NAD-dependent malate dehydrogenase complexed with NADP(H)
Biochem.Biophys.Res.Commun., 334, 2005
|
|
6LTB
 
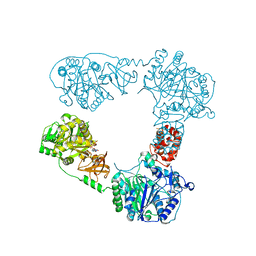 | |
6LTA
 
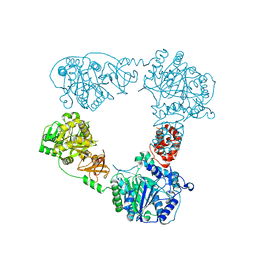 | |
6LTC
 
 | |
6LTD
 
 | |
1FLE
 
 | | CRYSTAL STRUCTURE OF ELAFIN COMPLEXED WITH PORCINE PANCREATIC ELASTASE | | Descriptor: | ELAFIN, ELASTASE | | Authors: | Tsunemi, M, Matsuura, Y, Sakakibara, S, Katsube, Y. | | Deposit date: | 1996-07-04 | | Release date: | 1997-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an elastase-specific inhibitor elafin complexed with porcine pancreatic elastase determined at 1.9 A resolution.
Biochemistry, 35, 1996
|
|
8X6R
 
 | | KRasG12C in complex with inhibitor | | Descriptor: | 1-[7-[6-ethenyl-8-ethoxy-7-(5-methyl-1~{H}-indazol-4-yl)-2-(1-methylpiperidin-4-yl)oxy-quinazolin-4-yl]-2,7-diazaspiro[3.5]nonan-2-yl]propan-1-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Amano, Y, Tateishi, Y. | | Deposit date: | 2023-11-21 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of ASP6918, a KRAS G12C inhibitor: Synthesis and structure-activity relationships of 1-{2,7-diazaspiro[3.5]non-2-yl}prop-2-en-1-one derivatives as covalent inhibitors with good potency and oral activity for the treatment of solid tumors.
Bioorg.Med.Chem., 98, 2023
|
|
6DL7
 
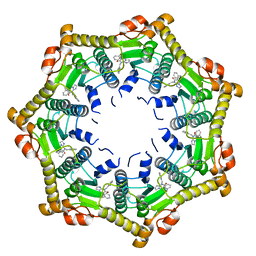 | | Human mitochondrial ClpP in complex with ONC201 (TIC10) | | Descriptor: | 7-benzyl-4-[(2-methylphenyl)methyl]-6,7,8,9-tetrahydroimidazo[1,2-a]pyrido[3,4-e]pyrimidin-5(4H)-one, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Halgas, O, Zarabi, S.F, Schimmer, A, Pai, E.F. | | Deposit date: | 2018-05-31 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mitochondrial ClpP-Mediated Proteolysis Induces Selective Cancer Cell Lethality.
Cancer Cell, 35, 2019
|
|
1L3H
 
 | | NMR structure of P41icf, a potent inhibitor of human cathepsin L | | Descriptor: | MHC CLASS II-ASSOCIATED P41 INVARIANT CHAIN FRAGMENT (P41icf) | | Authors: | Chiva, C, Barthe, P, Codina, A, Giralt, E. | | Deposit date: | 2002-02-27 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Synthesis and NMR structure of P41ICF, a potent inhibitor of human cathepsin L
J.Am.Chem.Soc., 125, 2003
|
|
1RSM
 
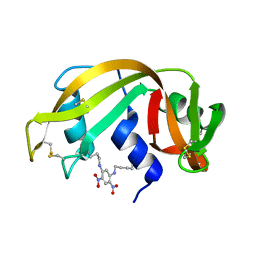 | | THE 2-ANGSTROMS RESOLUTION STRUCTURE OF A THERMOSTABLE RIBONUCLEASE A CHEMICALLY CROSS-LINKED BETWEEN LYSINE RESIDUES 7 AND 41 | | Descriptor: | DINITROPHENYLENE, RIBONUCLEASE A | | Authors: | Weber, P.C, Sheriff, S, Ohlendorf, D.H, Finzel, B.C, Salemme, F.R. | | Deposit date: | 1985-08-27 | | Release date: | 1986-01-21 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2-A resolution structure of a thermostable ribonuclease A chemically cross-linked between lysine residues 7 and 41.
Proc.Natl.Acad.Sci.USA, 82, 1985
|
|
5GSE
 
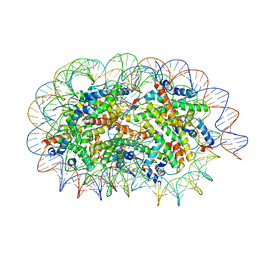 | | Crystal structure of unusual nucleosome | | Descriptor: | DNA (250-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kato, D, Osakabe, A, Arimura, Y, Park, S.Y, Kurumizaka, H. | | Deposit date: | 2016-08-16 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structure of the overlapping dinucleosome composed of hexasome and octasome
Science, 356, 2017
|
|
5B3V
 
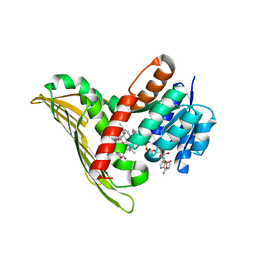 | |
5B3U
 
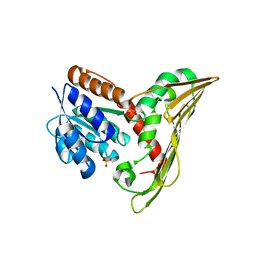 | |
5B3T
 
 | |
4ZY3
 
 | | Crystal Structure of Keap1 in Complex with a small chemical compound, K67 | | Descriptor: | FORMIC ACID, Kelch-like ECH-associated protein 1, N,N'-[2-(2-oxopropyl)naphthalene-1,4-diyl]bis(4-ethoxybenzenesulfonamide) | | Authors: | Fukutomi, T, Iso, T, Suzuki, T, Takagi, K, Mizushima, T, Komatsu, M, Yamamoto, M. | | Deposit date: | 2015-05-21 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming
Nat Commun, 7, 2016
|
|
1HBT
 
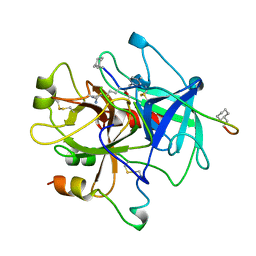 | |
1MLD
 
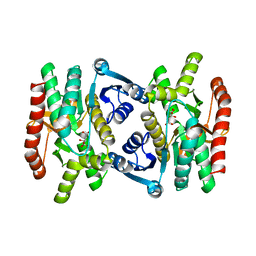 | | REFINED STRUCTURE OF MITOCHONDRIAL MALATE DEHYDROGENASE FROM PORCINE HEART AND THE CONSENSUS STRUCTURE FOR DICARBOXYLIC ACID OXIDOREDUCTASES | | Descriptor: | CITRIC ACID, MALATE DEHYDROGENASE | | Authors: | Gleason, W.B, Fu, Z, Birktoft, J.J, Banaszak, L.J. | | Deposit date: | 1994-01-24 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Refined crystal structure of mitochondrial malate dehydrogenase from porcine heart and the consensus structure for dicarboxylic acid oxidoreductases.
Biochemistry, 33, 1994
|
|
7DVS
 
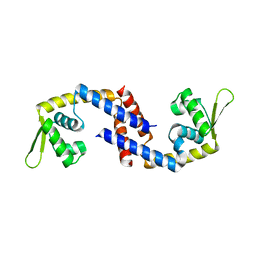 | | Crystal structure of Apo (heme-free) PefR | | Descriptor: | MarR family transcriptional regulator | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival.
Commun Biol, 4, 2021
|
|
1IS2
 
 | |
7CUF
 
 | |
