4U5F
 
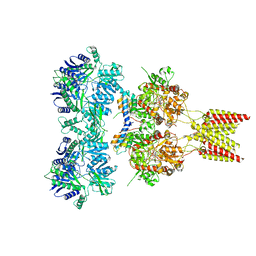 | | Crystal structure of GluA2, con-ikot-ikot snail toxin, partial agonist KA and postitive modulator (R,R)-2b complex, GluA2cryst2 construct | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Con-ikot-ikot, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-24 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism.
Science, 345, 2014
|
|
4U5C
 
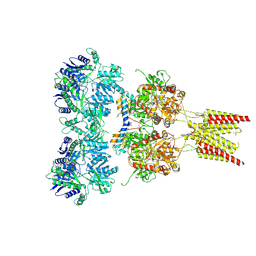 | | Crystal structure of GluA2, con-ikot-ikot snail toxin, partial agonist FW and postitive modulator (R,R)-2b complex | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Con-ikot-ikot, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.6883 Å) | | Cite: | X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism.
Science, 345, 2014
|
|
4U5E
 
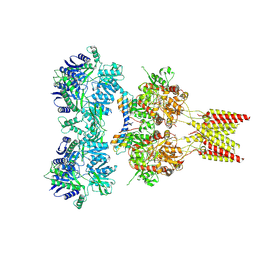 | | Crystal structure of GluA2 T625G, con-ikot-ikot snail toxin, partial agonist KA and postitive modulator (R,R)-2b complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Con-ikot-ikot, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5073 Å) | | Cite: | X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism.
Science, 345, 2014
|
|
4MM4
 
 | |
4MM8
 
 | |
4MMF
 
 | | Crystal structure of LeuBAT (delta5 mutant) in complex with mazindol | | Descriptor: | (5R)-5-(4-chlorophenyl)-2,5-dihydro-3H-imidazo[2,1-a]isoindol-5-ol, SODIUM ION, Transporter, ... | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
4NTW
 
 | | Structure of acid-sensing ion channel in complex with snake toxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, Basic phospholipase A2 homolog Tx-beta, ... | | Authors: | Baconguis, I, Bohlen, C.J, Goehring, A, Julius, D, Gouaux, E. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | X-ray structure of Acid-sensing ion channel 1-snake toxin complex reveals open state of a na(+)-selective channel.
Cell(Cambridge,Mass.), 156, 2014
|
|
4MM7
 
 | |
4MMC
 
 | |
4MMD
 
 | |
4NTX
 
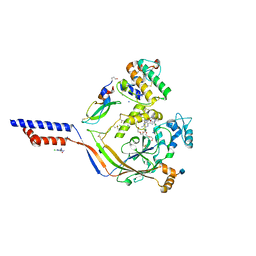 | | Structure of acid-sensing ion channel in complex with snake toxin and amiloride | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,5-DIAMINO-N-(AMINOIMINOMETHYL)-6-CHLOROPYRAZINECARBOXAMIDE, Acid-sensing ion channel 1, ... | | Authors: | Baconguis, I, Bohlen, C.J, Goehring, A, Julius, D, Gouaux, E. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | X-ray structure of Acid-sensing ion channel 1-snake toxin complex reveals open state of a na(+)-selective channel.
Cell(Cambridge,Mass.), 156, 2014
|
|
4NYK
 
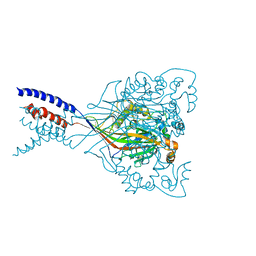 | | Structure of a membrane protein | | Descriptor: | Acid-sensing ion channel 1, CHLORIDE ION | | Authors: | Baconguis, I, Bohlen, C.J, Goehring, A, Julius, D, Gouaux, E. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Pore architecture and ion sites in acid-sensing ion channels and P2X receptors.
Nature, 460, 2009
|
|
4MM9
 
 | |
4MM5
 
 | | Crystal structure of LeuBAT (delta13 mutant) in complex with sertraline | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, SODIUM ION, Transporter | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
4MMB
 
 | | Crystal structure of LeuBAT (delta6 mutant) in complex with sertraline | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, SODIUM ION, Transporter, ... | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
4MMA
 
 | |
4MM6
 
 | |
4MME
 
 | | Crystal structure of LeuBAT (delta6 mutant) in complex with mazindol | | Descriptor: | (5R)-5-(4-chlorophenyl)-2,5-dihydro-3H-imidazo[2,1-a]isoindol-5-ol, SODIUM ION, Transporter, ... | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
4NTY
 
 | | Cesium sites in the crystal structure of acid-sensing ion channel in complex with snake toxin | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Acid-sensing ion channel 1, Basic phospholipase A2 homolog Tx-beta, ... | | Authors: | Baconguis, I, Bohlen, C.J, Goehring, A, Julius, D, Gouaux, E. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-19 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | X-ray structure of Acid-sensing ion channel 1-snake toxin complex reveals open state of a na(+)-selective channel.
Cell(Cambridge,Mass.), 156, 2014
|
|
1P1W
 
 | |
1P1Q
 
 | |
1P1U
 
 | |
1P1O
 
 | | Crystal structure of the GluR2 ligand-binding core (S1S2J) mutant L650T in complex with quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2, SULFATE ION | | Authors: | Armstrong, N, Mayer, M.L, Gouaux, E. | | Deposit date: | 2003-04-13 | | Release date: | 2003-06-10 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tuning activation of the AMPA-sensitive GluR2 ion channel by genetic
adjustment of agonist-induced conformational changes.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1P1N
 
 | |
1PB8
 
 | |
