1H0Z
 
 | | LEKTI domain six | | Descriptor: | SERINE PROTEASE INHIBITOR KAZAL-TYPE 5, CONTAINS HEMOFILTRATE PEPTIDE HF6478, HEMOFILTRATE PEPTIDE HF7665 | | Authors: | Lauber, T, Roesch, P, Marx, U.C. | | Deposit date: | 2002-07-01 | | Release date: | 2003-06-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Homologous Proteins with Different Folds: The Three-Dimensional Structures of Domains 1 and 6 of the Multiple Kazal-Type Inhibitor Lekti
J.Mol.Biol., 328, 2003
|
|
4ZKE
 
 | |
4ZKD
 
 | |
5A13
 
 | |
5A12
 
 | |
5CAW
 
 | |
5C9V
 
 | | Structure of human Parkin G319A | | Descriptor: | E3 ubiquitin-protein ligase parkin, GLYCEROL, SULFATE ION, ... | | Authors: | Wauer, T, Komander, D. | | Deposit date: | 2015-06-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of phospho-ubiquitin-induced PARKIN activation.
Nature, 524, 2015
|
|
4P5R
 
 | | Structure of oxidized W45Y mutant of amicyanin | | Descriptor: | Amicyanin, COPPER (II) ION, SODIUM ION | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | The sole tryptophan of amicyanin enhances its thermal stability but does not influence the electronic properties of the type 1 copper site.
Arch.Biochem.Biophys., 550-551, 2014
|
|
8OFE
 
 | | E.coli Peptide Deformylase with bound inhibitor | | Descriptor: | (2R)-2-[[methanoyl(oxidanyl)amino]methyl]-N-[(2S)-3-methyl-1-oxidanylidene-1-[2-(sulfanylmethyl)pyrrolidin-1-yl]butan-2-yl]heptanamide, Peptide deformylase, ZINC ION | | Authors: | Kirschner, H, Stoll, R, Hofmann, E. | | Deposit date: | 2023-03-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Insights into Antibacterial Payload Release from Gold Nanoparticles Bound to E. coli Peptide Deformylase.
Chemmedchem, 19, 2024
|
|
4P5S
 
 | | Structure of reduced W45Y mutant of amicyanin | | Descriptor: | Amicyanin, COPPER (I) ION | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The sole tryptophan of amicyanin enhances its thermal stability but does not influence the electronic properties of the type 1 copper site.
Arch.Biochem.Biophys., 550-551, 2014
|
|
4MOT
 
 | | Structure of Streptococcus pneumonia pare in complex with AZ13072886 | | Descriptor: | 1-[4-(3-methylbutyl)-5-oxo-6-(pyridin-3-yl)-4,5-dihydro[1,3]thiazolo[5,4-b]pyridin-2-yl]-3-prop-2-en-1-ylurea, Topoisomerase IV subunit B | | Authors: | Ogg, D, Boriack-Sjodin, P.A. | | Deposit date: | 2013-09-12 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thiazolopyridone ureas as DNA gyrase B inhibitors: Optimization of antitubercular activity and efficacy.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
1J1U
 
 | | Crystal structure of archaeal tyrosyl-tRNA synthetase complexed with tRNA(Tyr) and L-tyrosine | | Descriptor: | MAGNESIUM ION, TYROSINE, Tyrosyl-tRNA synthetase, ... | | Authors: | Kobayashi, T, Nureki, O, Ishitani, R, Tukalo, M, Cusack, S, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-17 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion
NAT.STRUCT.BIOL., 10, 2003
|
|
2W55
 
 | | Crystal Structure of Xanthine Dehydrogenase (E232Q variant) from Rhodobacter capsulatus in Complex with Hypoxanthine | | Descriptor: | BARIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Doebbler, J.A, Truglio, J.J, Leimkuhler, S, Kisker, C. | | Deposit date: | 2008-12-04 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mechanism of Substrate and Inhibitor Binding of Rhodobacter Capsulatus Xanthine Dehydrogenase.
J.Biol.Chem., 284, 2009
|
|
2W54
 
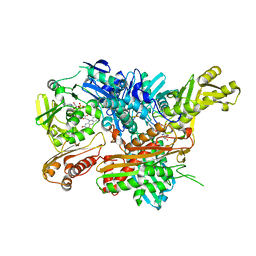 | | Crystal Structure of Xanthine Dehydrogenase from Rhodobacter capsulatus in Complex with Bound Inhibitor Pterin-6-aldehyde | | Descriptor: | 6-HYDROXYMETHYLPTERIN, BARIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Doebbler, J.A, Truglio, J.J, Leimkuhler, S, Kisker, C. | | Deposit date: | 2008-12-04 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mechanism of Substrate and Inhibitor Binding of Rhodobacter Capsulatus Xanthine Dehydrogenase.
J.Biol.Chem., 284, 2009
|
|
2W2H
 
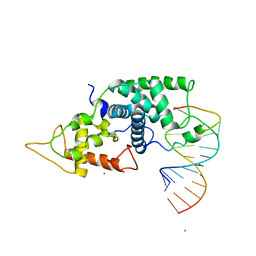 | |
3T3C
 
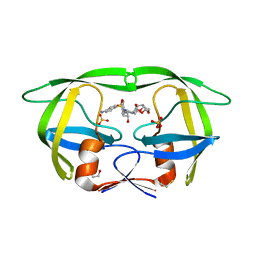 | | Structure of HIV PR resistant patient derived mutant (comprising 22 mutations) in complex with DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, BETA-MERCAPTOETHANOL, HIV-1 protease, ... | | Authors: | Rezacova, P, Kozisek, M, Konvalinka, J, Saskova, K.G. | | Deposit date: | 2011-07-25 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations in HIV-1 gag and pol Compensate for the Loss of Viral Fitness Caused by a Highly Mutated Protease.
Antimicrob.Agents Chemother., 56, 2012
|
|
2W3S
 
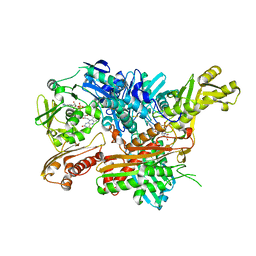 | | Crystal Structure of Xanthine Dehydrogenase (desulfo form) from Rhodobacter capsulatus in Complex with Xanthine | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dietzel, U, Kuper, J, Leimkuhler, S, Kisker, C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of Substrate and Inhibitor Binding of Rhodobacter Capsulatus Xanthine Dehydrogenase.
J.Biol.Chem., 284, 2009
|
|
2W3R
 
 | | Crystal Structure of Xanthine Dehydrogenase (desulfo form) from Rhodobacter capsulatus in complex with hypoxanthine | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dietzel, U, Kuper, J, Leimkuhler, S, Kisker, C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of Substrate and Inhibitor Binding of Rhodobacter Capsulatus Xanthine Dehydrogenase.
J.Biol.Chem., 284, 2009
|
|
5OR2
 
 | | Crystal structures of PYR1/HAB1 in complex with synthetic analogues of Abscisic Acid | | Descriptor: | (2~{Z},4~{E})-3-cyclopropyl-5-[(1~{S})-2,6,6-trimethyl-1-oxidanyl-4-oxidanylidene-cyclohex-2-en-1-yl]penta-2,4-dienoic acid, Abscisic acid receptor PYR1, MANGANESE (II) ION, ... | | Authors: | Freigang, J. | | Deposit date: | 2017-08-15 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the in Vitro and in Vivo SAR of Abscisic Acid - Exploring Unprecedented Variations of the Side Chain via Cross-Coupling-Mediated Syntheses
Eur.J.Org.Chem., 2018
|
|
6ST6
 
 | | Crystal Structure of Domain Swapped Trp Repressor V58I Variant | | Descriptor: | ISOPROPYL ALCOHOL, Trp operon repressor | | Authors: | Sprenger, J, Lawson, C.L, Carey, J, Drouard, F, von Wachenfeldt, C, Schulz, A, Linse, S, Lo Leggio, L. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of Val58Ile tryptophan repressor in a domain-swapped array in the presence and absence of L-tryptophan.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
3ZM3
 
 | | Catalytic domain of human SHP2 | | Descriptor: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 11 | | Authors: | Bohm, K, Schuetz, A, Roske, Y, Heinemann, U. | | Deposit date: | 2013-02-04 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selective Inhibitors of the Protein Tyrosine Phosphatase Shp2 Block Cellular Motility and Growth of Cancer Cells in Vitro and in Vivo.
Chemmedchem, 10, 2015
|
|
3ZM0
 
 | | Catalytic domain of human SHP2 | | Descriptor: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 11 | | Authors: | Bohm, K, Schuetz, A, Roske, Y, Heinemann, U. | | Deposit date: | 2013-02-04 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selective Inhibitors of the Protein Tyrosine Phosphatase Shp2 Block Cellular Motility and Growth of Cancer Cells in Vitro and in Vivo.
Chemmedchem, 10, 2015
|
|
3ZM2
 
 | | Catalytic domain of human SHP2 | | Descriptor: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 11 | | Authors: | Bohm, K, Schuetz, A, Roske, Y, Heinemann, U. | | Deposit date: | 2013-02-04 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selective Inhibitors of the Protein Tyrosine Phosphatase Shp2 Block Cellular Motility and Growth of Cancer Cells in Vitro and in Vivo.
Chemmedchem, 10, 2015
|
|
6ST7
 
 | | Crystal Structure of Domain Swapped Trp Repressor V58I Variant with bound L-trp | | Descriptor: | ISOPROPYL ALCOHOL, TRYPTOPHAN, Trp operon repressor | | Authors: | Sprenger, J, Lawson, C.L, Carey, J, Drouard, F, von Wachenfeldt, C, Schulz, A, Linse, S, Lo Leggio, L. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of Val58Ile tryptophan repressor in a domain-swapped array in the presence and absence of L-tryptophan.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
3ZM1
 
 | | Catalytic domain of human SHP2 | | Descriptor: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 11 | | Authors: | Bohm, K, Schuetz, A, Roske, Y, Heinemann, U. | | Deposit date: | 2013-02-04 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective Inhibitors of the Protein Tyrosine Phosphatase Shp2 Block Cellular Motility and Growth of Cancer Cells in Vitro and in Vivo.
Chemmedchem, 10, 2015
|
|
