8T7C
 
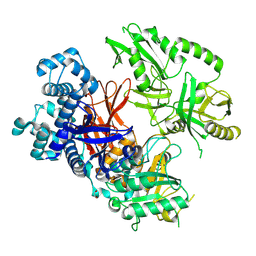 | | Crystal structure of human phospholipase C gamma 2 | | Descriptor: | 1,2-ETHANEDIOL, 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-2, CALCIUM ION | | Authors: | Chen, Y, Choi, H, Zhuang, N, Hu, L, Qian, D, Wang, J. | | Deposit date: | 2023-06-20 | | Release date: | 2024-06-26 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal and cryo-EM structures of PLC gamma 2 reveal dynamic interdomain recognitions in autoinhibition.
Sci Adv, 10, 2024
|
|
8Y0D
 
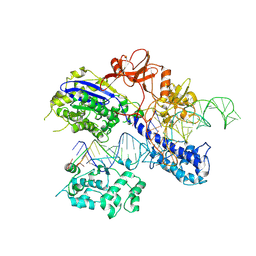 | |
8J54
 
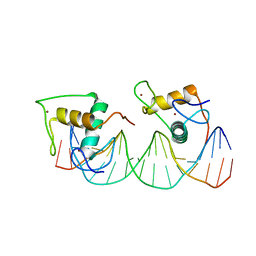 | | Crystal structure of RXR/DR2 complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*CP*CP*TP*AP*CP*TP*GP*AP*CP*CP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*GP*GP*TP*CP*AP*GP*TP*AP*GP*GP*TP*CP*AP*TP*G)-3'), Retinoic acid receptor RXR, ... | | Authors: | Chen, Y, Jiang, L. | | Deposit date: | 2023-04-21 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural characterization of the DNA binding mechanism of retinoic acid-related orphan receptor gamma.
Structure, 32, 2024
|
|
7XY7
 
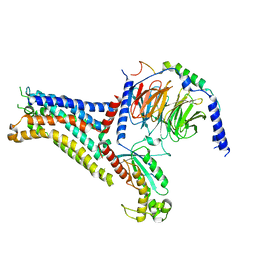 | | Adenosine receptor bound to a non-selective agonist in complex with a G protein obtained by cryo-EM | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Zhang, J.Y, Chen, Y, Hua, T, Song, G.J. | | Deposit date: | 2022-05-31 | | Release date: | 2023-05-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Cryo-EM structure of the human adenosine A 2B receptor-G s signaling complex.
Sci Adv, 8, 2022
|
|
7XY6
 
 | | Adenosine receptor bound to an agonist in complex with G protein obtained by cryo-EM | | Descriptor: | 2-[6-azanyl-3,5-dicyano-4-[4-(cyclopropylmethoxy)phenyl]pyridin-2-yl]sulfanylethanamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, J.Y, Chen, Y, Hua, T, Song, G.J. | | Deposit date: | 2022-05-31 | | Release date: | 2023-05-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM structure of the human adenosine A 2B receptor-G s signaling complex.
Sci Adv, 8, 2022
|
|
8KGO
 
 | |
7YOT
 
 | |
7YOU
 
 | |
7YOV
 
 | |
7YA2
 
 | |
5YC2
 
 | |
5YCA
 
 | | Crystal structure of inner membrane protein Bqt4 in complex with LEM2 | | Descriptor: | Lap-Emerin-Man domain protein 2, Ubiquitin-like protein SMT3,Bouquet formation protein 4 | | Authors: | Chen, Y, Hu, C. | | Deposit date: | 2017-09-07 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural insights into chromosome attachment to the nuclear envelope by an inner nuclear membrane protein Bqt4 in fission yeast.
Nucleic Acids Res., 47, 2019
|
|
6A6W
 
 | |
8G6U
 
 | | Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer with LMHS mutations in complex with 8ANC195 and 10-1074 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CRF-1_AE T/F100 HIV-1 gp41, ... | | Authors: | Chen, Y, Zhou, F, Huang, R, Tolbert, W, Pazgier, M. | | Deposit date: | 2023-02-16 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Nat Commun, 14, 2023
|
|
6J0Q
 
 | | Crystal structure of P domain from GII.11 swine norovirus | | Descriptor: | VP1 capsid protein | | Authors: | Chen, Y. | | Deposit date: | 2018-12-25 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
7X4C
 
 | | Native CD-NTase EfCdnE | | Descriptor: | EfCdnE, L(+)-TARTARIC ACID | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
7X4P
 
 | | CD-NTase EfCdnE in complex with intermediate pppUpU | | Descriptor: | EfCdnE, MAGNESIUM ION, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-03 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
7X4A
 
 | | Native CD-NTase ClCdnE | | Descriptor: | ClCdnE | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
7X4F
 
 | | Native CD-NTase LpCdnE | | Descriptor: | Cyclic dipyrimidine nucleotide synthase, SULFATE ION | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
7X4Q
 
 | | LpCdnE UTP Mg complex | | Descriptor: | Cyclic dipyrimidine nucleotide synthase, MAGNESIUM ION, URIDINE 5'-TRIPHOSPHATE | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
7X4G
 
 | | CD-NTase ClCdnE in complex with substrate UTP | | Descriptor: | ClCdnE, MAGNESIUM ION, URIDINE 5'-TRIPHOSPHATE | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
7X4T
 
 | | LpCdnE UMPNPP Mg complex | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, Cyclic dipyrimidine nucleotide synthase, GLYCEROL, ... | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
7XVN
 
 | | Structural basis for DNA recognition feature of retinoid-related orphan receptors | | Descriptor: | DNA (5'-D(P*CP*AP*TP*GP*AP*CP*CP*TP*AP*CP*TP*GP*AP*CP*CP*TP*AP*G)-3'), DNA (5'-D(P*CP*TP*AP*GP*GP*TP*CP*AP*GP*TP*AP*GP*GP*TP*CP*AP*TP*G)-3'), Nuclear receptor ROR-gamma, ... | | Authors: | Chen, Y, Jiang, L. | | Deposit date: | 2022-05-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural characterization of the DNA binding mechanism of retinoic acid-related orphan receptor gamma.
Structure, 32, 2024
|
|
5Y18
 
 | |
5ZV7
 
 | | P domain of GII.17-2014/15 complexed with B-trisaccharide | | Descriptor: | VP1, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
