6RFX
 
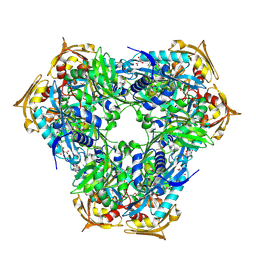 | | Crystal structure of Eis2 from Mycobacterium abscessus | | Descriptor: | ACETATE ION, CITRIC ACID, Eis2, ... | | Authors: | Blaise, M, Kremer, L, Olieric, V, Alsarraf, H, Ung, K.L. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-10 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the aminoglycosides N-acetyltransferase Eis2 from Mycobacterium abscessus.
Febs J., 286, 2019
|
|
6RFY
 
 | | Crystal structure of Eis2 form Mycobacterium abscessus | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Eis2, SULFATE ION | | Authors: | Blaise, M, Kremer, L, Olieric, V, Alsarraf, H, Ung, K.L. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the aminoglycosides N-acetyltransferase Eis2 from Mycobacterium abscessus.
Febs J., 286, 2019
|
|
6F7C
 
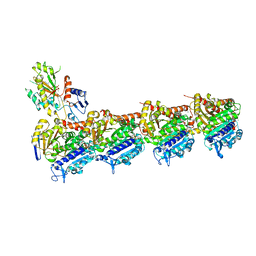 | | TUBULIN-Compound 12 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,4,5-trimethoxy-~{N}-[(~{E})-naphthalen-1-ylmethylideneamino]benzamide, CALCIUM ION, ... | | Authors: | Muehlethaler, T, Prota, A.E, Steinmetz, M.O. | | Deposit date: | 2017-12-08 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structural Basis of Colchicine-Site targeting Acylhydrazones active against Multidrug-Resistant Acute Lymphoblastic Leukemia.
Iscience, 21, 2019
|
|
7ANQ
 
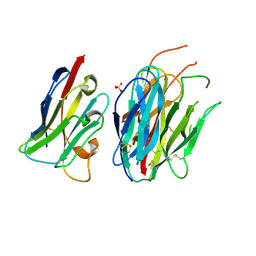 | | Complete PCSK9 C-ter domain in complex with VHH P1.40 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Proprotein convertase subtilisin/kexin type 9, SULFATE ION, ... | | Authors: | Ciccone, L, Legrand, P, Stura, E.A, Dive, V, Seidahn, N.G, Fruchart Gaillard, C. | | Deposit date: | 2020-10-12 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular interactions of PCSK9 with an inhibitory nanobody, CAP1 and HLA-C: Functional regulation of LDLR levels.
Mol Metab, 67, 2022
|
|
8JZM
 
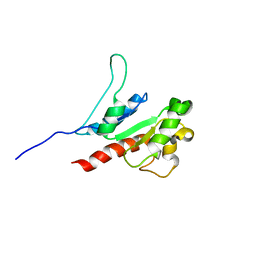 | | The inhibitor of Toll-like receptor signaling o-vanillin binds covalently to MAL/TIRAP Lys-210 | | Descriptor: | Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Rahaman, M.H, Jia, X, Maxwell, M.J, Mobli, M, Kobe, B. | | Deposit date: | 2023-07-05 | | Release date: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | o-Vanillin binds covalently to MAL/TIRAP Lys-210 but independently inhibits TLR2.
J Enzyme Inhib Med Chem, 39, 2024
|
|
7AGL
 
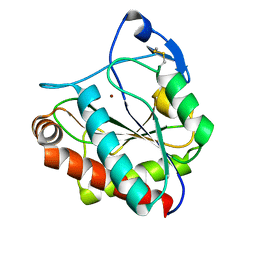 | |
7AGO
 
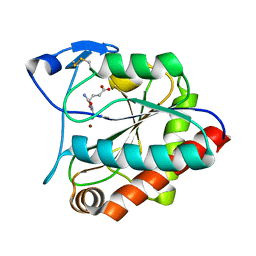 | | crystal structure of the N-acetylmuramyl-L-alanine amidase, Ami1, from Mycobacterium abscessus bound to L-Alanine-D-isoglutamine | | Descriptor: | ALANINE, D-alpha-glutamine, N-acetylmuramoyl-L-alanine amidase, ... | | Authors: | Blaise, M. | | Deposit date: | 2020-09-23 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional Characterization of the N -Acetylmuramyl-l-Alanine Amidase, Ami1, from Mycobacterium abscessus .
Cells, 9, 2020
|
|
5MDI
 
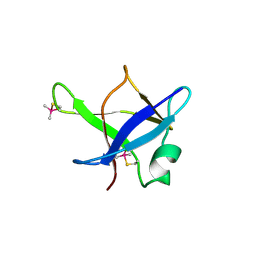 | | Crystal structure of TDP-43 N-terminal domain at 2.1 A resolution | | Descriptor: | ACETATE ION, TAR DNA-binding protein 43 | | Authors: | Afroz, T, Hock, E.-M, Ernst, P, Foglieni, C, Jambeau, M, Gilhespy, L, Laferriere, F, Maniecka, Z, Plueckthun, A, Mittl, P, Paganetti, P, Allain, F.H.T, Polymenidou, M. | | Deposit date: | 2016-11-11 | | Release date: | 2017-07-05 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation.
Nat Commun, 8, 2017
|
|
5ZOB
 
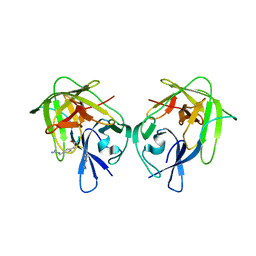 | |
5ZMS
 
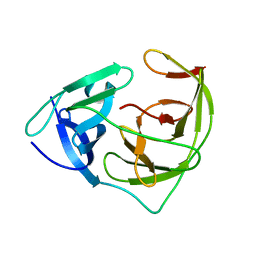 | |
5ZMQ
 
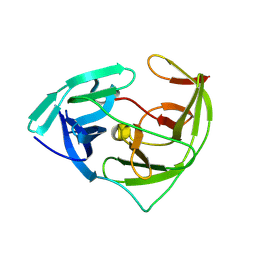 | |
5ZXN
 
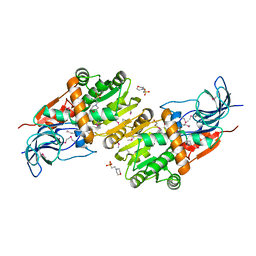 | | Crystal structure of CurA from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NADP-dependent oxidoreductase | | Authors: | Kim, M.-K, Bae, D.-W, Cha, S.-S. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | Structural and Biochemical Characterization of the Curcumin-Reducing Activity of CurA from Vibrio vulnificus.
J. Agric. Food Chem., 66, 2018
|
|
5ZXU
 
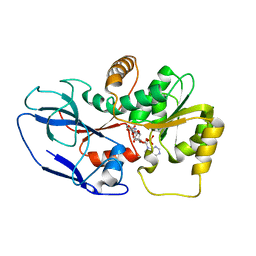 | |
5V4L
 
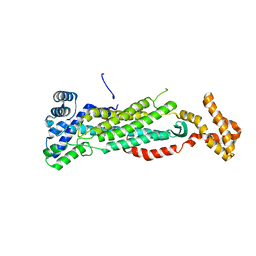 | |
1HUP
 
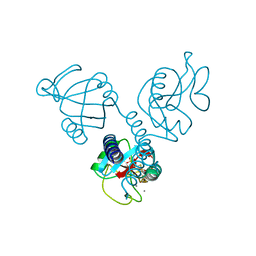 | |
7RZ3
 
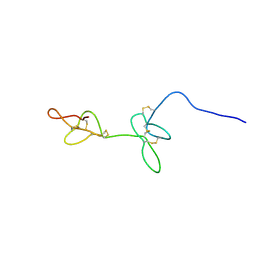 | |
6T84
 
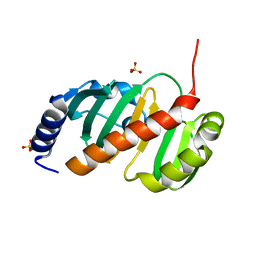 | |
5EA9
 
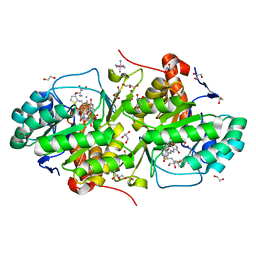 | | Crystal Structure of Trypanosoma cruzi Dihydroorotate Dehydrogenase in Complex with Neq0130 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(E)-3-thiophen-2-ylprop-2-enylidene]-1,3-diazinane-2,4,6-trione, COBALT HEXAMMINE(III), ... | | Authors: | Rocha, J.R, Inaoka, D.K, Cheleski, J, Shiba, T, Harada, S, Montanari, C.A, Kita, K. | | Deposit date: | 2015-10-15 | | Release date: | 2016-10-19 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Exploring Trypanosoma cruzi Dihydroorotate Dehydrogenase Active Site Plasticity for the Discovery of Potent and Selective Inhibitors with Trypanocidal Activity
To be Published
|
|
2N6N
 
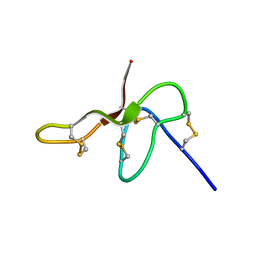 | | Structure Determination for spider toxin, U4-agatoxin-Ao1a | | Descriptor: | U4-agatoxin-Ao1a | | Authors: | Pineda, S.S, Chin, Y.K.-Y, Mobli, M.S, King, G.F. | | Deposit date: | 2015-08-27 | | Release date: | 2016-08-31 | | Last modified: | 2023-03-08 | | Method: | SOLUTION NMR | | Cite: | Structural venomics reveals evolution of a complex venom by duplication and diversification of an ancient peptide-encoding gene.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2MUN
 
 | |
2N8K
 
 | | Chemical Shift Assignments and Structure Determination for spider toxin, U33-theraphotoxin-Cg1c | | Descriptor: | U33-theraphotoxin-Cg1b | | Authors: | Chin, Y.K.-Y, Pineda, S.S, Mobli, M, King, G.F. | | Deposit date: | 2015-10-20 | | Release date: | 2016-08-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural venomics reveals evolution of a complex venom by duplication and diversification of an ancient peptide-encoding gene.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2N6R
 
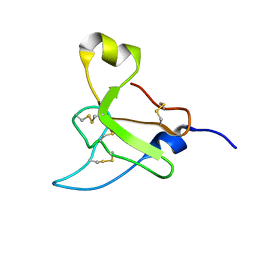 | |
8SJV
 
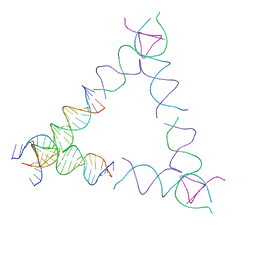 | | [4T24] Self-assembling left-handed tensegrity triangle with 24 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(P*CP*TP*TP*GP*TP*AP*GP*TP*CP*TP*CP*AP*CP*CP*AP*CP*TP*GP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*GP*AP*AP*CP*AP*CP*TP*CP*CP*TP*GP*AP*GP*AP*CP*TP*AP*CP*AP*A)-3'), DNA (5'-D(P*GP*AP*CP*AP*TP*CP*AP*CP*AP*GP*TP*GP*GP*AP*CP*TP*AP*CP*AP*AP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (8.59 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJR
 
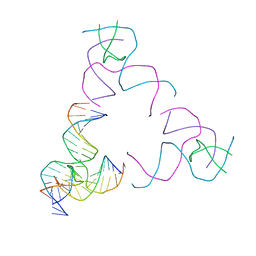 | | [3T17] Self-assembling right-handed tensegrity triangle with 17 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*AP*GP*CP*CP*TP*GP*AP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*CP*TP*GP*TP*GP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*GP*TP*AP*TP*TP*CP*AP*CP*CP*AP*CP*GP*AP*T)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (5.25 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJM
 
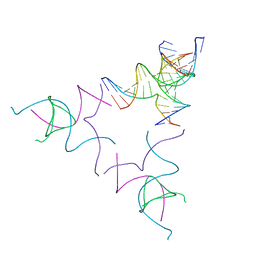 | | [3T12] Self-assembling left-handed tensegrity triangle with 12 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*AP*TP*CP*GP*CP*CP*TP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*CP*GP*CP*AP*TP*GP*TP*GP*GP*CP*GP*AP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*AP*CP*AP*TP*GP*CP*GP*AP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (8.08 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
