1EG4
 
 | | STRUCTURE OF A DYSTROPHIN WW DOMAIN FRAGMENT IN COMPLEX WITH A BETA-DYSTROGLYCAN PEPTIDE | | Descriptor: | BETA-DYSTROGLYCAN, DYSTROPHIN | | Authors: | Huang, X, Poy, F, Zhang, R, Joachimiak, A, Sudol, M, Eck, M.J. | | Deposit date: | 2000-02-11 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan.
Nat.Struct.Biol., 7, 2000
|
|
1MQA
 
 | | Crystal structure of high affinity alphaL I domain in the absence of ligand or metal | | Descriptor: | Integrin alpha-L | | Authors: | Shimaoka, T, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, Zhang, R, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1MQ9
 
 | | Crystal structure of high affinity alphaL I domain with ligand mimetic crystal contact | | Descriptor: | Integrin alpha-L, MANGANESE (II) ION | | Authors: | Shimaoka, M, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, McCormack, A, Zhang, R, Joachimiak, A, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1MBJ
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBK
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBE
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBH
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 2 | | Descriptor: | C-MYB | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-09-15 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBG
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 2 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
3DOB
 
 | | Peptide-binding domain of Heat shock 70 kDa protein F44E5.5 from C.elegans. | | Descriptor: | BETA-MERCAPTOETHANOL, Heat shock 70 kDa protein F44E5.5 | | Authors: | Osipiuk, J, Hatzos, C, Gu, M, Zhang, R, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-03 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | X-ray crystal structure of Peptide-binding domain of Heat shock 70 kDa protein F44E5.5 from C.elegans.
To be Published
|
|
7SJ8
 
 | | 13pf wildtype microtubule from recombinant human tubulin decorated with kinesin | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | LaFrance, B.J, Greber, B.J, Zhang, R, McCollum, C, Nogales, E. | | Deposit date: | 2021-10-16 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural transitions in the GTP cap visualized by cryo-electron microscopy of catalytically inactive microtubules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SJ9
 
 | | 13pf E254A microtubule from recombinant human tubulin decorated with EB3 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Microtubule-associated protein RP/EB family member 3, ... | | Authors: | LaFrance, B.J, Greber, B.J, Zhang, R, McCollum, C, Nogales, E. | | Deposit date: | 2021-10-16 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural transitions in the GTP cap visualized by cryo-electron microscopy of catalytically inactive microtubules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SJA
 
 | | Undecorated 13pf E254N microtubule from recombinant human tubulin | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Tubulin alpha-1B chain, ... | | Authors: | LaFrance, B.J, Greber, B.J, Zhang, R, McCollum, C, Nogales, E. | | Deposit date: | 2021-10-16 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural transitions in the GTP cap visualized by cryo-electron microscopy of catalytically inactive microtubules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SJ7
 
 | | Undecorated 13pf wildtype microtubule from recombinant human tubulin | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | LaFrance, B.J, Greber, B.J, Zhang, R, McCollum, C, Nogales, E. | | Deposit date: | 2021-10-16 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural transitions in the GTP cap visualized by cryo-electron microscopy of catalytically inactive microtubules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2G5C
 
 | | Crystal Structure of Prephenate Dehydrogenase from Aquifex aeolicus | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, prephenate dehydrogenase | | Authors: | Sun, W, Singh, S, Zhang, R, Turnbull, J.L, Christendat, D. | | Deposit date: | 2006-02-22 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Prephenate Dehydrogenase from Aquifex aeolicus: Insights into the Catalytic Mechanism
J.Biol.Chem., 281, 2006
|
|
5W3J
 
 | | Yeast microtubule stabilized with Taxol assembled from mutated tubulin | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Howes, S.C, Geyer, E.A, LaFrance, B, Zhang, R, Kellogg, E.H, Westermann, S, Rice, L.M, Nogales, E. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural differences between yeast and mammalian microtubules revealed by cryo-EM.
J. Cell Biol., 216, 2017
|
|
4GHT
 
 | | Crystal structure of EV71 3C proteinase in complex with AG7088 | | Descriptor: | 3C proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5WTL
 
 | | Crystal structure of the periplasmic portion of outer membrane protein A (OmpA) from Capnocytophaga gingivalis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, OmpA family protein, ... | | Authors: | Dai, S, Tan, K, Ye, S, Zhang, R. | | Deposit date: | 2016-12-13 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Structure of thrombospondin type 3 repeats in bacterial outer membrane protein A reveals its intra-repeat disulfide bond-dependent calcium-binding capability.
Cell Calcium, 66, 2017
|
|
5WTP
 
 | | Crystal structure of the C-terminal domain of outer membrane protein A (OmpA) from Capnocytophaga gingivalis | | Descriptor: | OmpA family protein, SULFATE ION | | Authors: | Dai, S, Tan, K, Ye, S, Zhang, R. | | Deposit date: | 2016-12-13 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of thrombospondin type 3 repeats in bacterial outer membrane protein A reveals its intra-repeat disulfide bond-dependent calcium-binding capability.
Cell Calcium, 66, 2017
|
|
1QK6
 
 | | Solution structure of huwentoxin-I by NMR | | Descriptor: | HUWENTOXIN-I | | Authors: | Qu, Y, Liang, S, Ding, J, Liu, X, Zhang, R, Gu, X. | | Deposit date: | 1999-07-10 | | Release date: | 1999-08-20 | | Last modified: | 2019-01-16 | | Method: | SOLUTION NMR | | Cite: | Proton Nuclear Magnetic Resonance Studies on Huwentoxin-I from the Venom of the Spider Selenocosmia Huwena:2.Three-Dimensional Structure in Solution
J.Protein Chem., 16, 1997
|
|
5CMZ
 
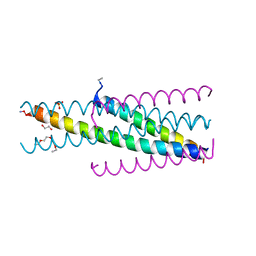 | | Artificial HIV fusion inhibitor AP3 fused to the C-terminus of gp41 NHR | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Artificial HIV entry inhibitor AP3, ... | | Authors: | Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Improved Pharmacological and Structural Properties of HIV Fusion Inhibitor AP3 over Enfuvirtide: Highlighting Advantages of Artificial Peptide Strategy.
Sci Rep, 5, 2015
|
|
5CN0
 
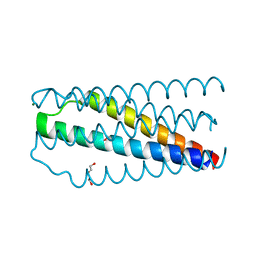 | | Artificial HIV fusion inhibitor AP2 fused to the C-terminus of gp41 NHR | | Descriptor: | DI(HYDROXYETHYL)ETHER, Envelope glycoprotein,AP2, MAGNESIUM ION | | Authors: | Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Improved Pharmacological and Structural Properties of HIV Fusion Inhibitor AP3 over Enfuvirtide: Highlighting Advantages of Artificial Peptide Strategy.
Sci Rep, 5, 2015
|
|
5CMU
 
 | |
1OXZ
 
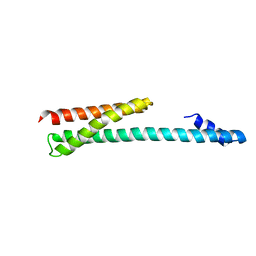 | | Crystal Structure of the Human GGA1 GAT domain | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Zhu, G, Zhai, P, He, X, Terzyan, S, Zhang, R, Joachimiak, A, Tang, J, Zhang, X.C. | | Deposit date: | 2003-04-03 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human GGA1 GAT Domain
Biochemistry, 42, 2003
|
|
6KZ1
 
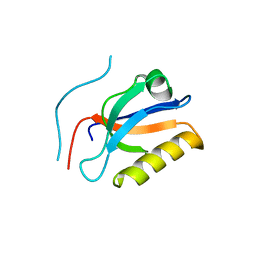 | | Complex structure of Whirlin and Myosin XVa | | Descriptor: | Myosin XVa, Whirlin | | Authors: | Lin, L, Wang, M, Shi, Y, Zhu, J, Zhang, R. | | Deposit date: | 2019-09-22 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Phase separation-mediated condensation of Whirlin-Myo15-Eps8 stereocilia tip complex.
Cell Rep, 34, 2021
|
|
5COA
 
 | | Crystal structure of iridoid synthase at 2.2-angstrom resolution | | Descriptor: | HEXAETHYLENE GLYCOL, Iridoid synthase, SULFATE ION | | Authors: | Qin, L, Zhu, Y, Ding, Z, Zhang, X, Ye, S, Zhang, R. | | Deposit date: | 2015-07-20 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of iridoid synthase in complex with NADP(+)/8-oxogeranial reveals the structural basis of its substrate specificity.
J.Struct.Biol., 194, 2016
|
|
