7AKC
 
 | | Structure of the of AcylTransferase domain of phenolphthiocerol/phtiocerol synthase A from Mycobacterium bovis (BCG) | | Descriptor: | Phenolpthiocerol synthesis type-I polyketide synthase ppsA, SODIUM ION | | Authors: | Brison, Y, Nahoum, V, Mourey, L, Maveyraud, L. | | Deposit date: | 2020-09-30 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Basis for Extender Unit Specificity of Mycobacterial Polyketide Synthases.
Acs Chem.Biol., 15, 2020
|
|
3K1Y
 
 | | X-ray structure of oxidoreductase from corynebacterium diphtheriae. orthorombic crystal form, northeast structural genomics consortium target cdr100d | | Descriptor: | SULFATE ION, oxidoreductase | | Authors: | Kuzin, A, Lew, S, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-29 | | Release date: | 2009-10-20 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Northeast Structural Genomics Consortium Target CdR100D
To be Published
|
|
3K3N
 
 | | Crystal structure of the catalytic core domain of human PHF8 | | Descriptor: | FE (II) ION, PHD finger protein 8 | | Authors: | Yu, L, Wang, Y, Huang, S, Wang, J, Deng, Z, Wu, W, Gong, W, Chen, Z. | | Deposit date: | 2009-10-03 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into a novel histone demethylase PHF8
Cell Res., 20, 2010
|
|
6ETY
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a inhibitor JHU3371 | | Descriptor: | (2~{S})-2-[[(2~{R})-4-methyl-1-oxidanyl-1-oxidanylidene-pentan-2-yl]carbamoyloxy]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Novakova, Z, Motlova, L. | | Deposit date: | 2017-10-27 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and computational basis for potent inhibition of glutamate carboxypeptidase II by carbamate-based inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
3JA4
 
 | |
7AGU
 
 | | Structure of the S726F mutant of AcylTransferase domain of Mycocerosic Acid Synthase from Mycobacterium tuberculosis acylated with MethylMalonyl-coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, METHYLMALONIC ACID, Mycocerosic acid synthase | | Authors: | Brison, Y, Mourey, L, Maveyraud, L. | | Deposit date: | 2020-09-23 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Basis for Extender Unit Specificity of Mycobacterial Polyketide Synthases.
Acs Chem.Biol., 15, 2020
|
|
3JC7
 
 | | Structure of the eukaryotic replicative CMG helicase and pumpjack motion | | Descriptor: | Cell division control protein 45, DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, ... | | Authors: | Li, H, Bai, L, Yuan, Z, Sun, J, Georgescu, R.E, Liu, J, O'Donnell, M.E. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3J6Q
 
 | | Identification of the active sites in the methyltransferases of a transcribing dsRNA virus | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, Structural protein VP3 | | Authors: | Zhu, B, Yang, C, Liu, H, Cheng, L, Song, F, Zeng, S, Huang, X, Ji, G, Zhu, P. | | Deposit date: | 2014-03-20 | | Release date: | 2014-10-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Identification of the active sites in the methyltransferases of a transcribing dsRNA virus.
J.Mol.Biol., 426, 2014
|
|
7AGT
 
 | |
3JSR
 
 | | X-Ray structure of All0216 protein from Nostoc sp. PCC 7120 at the resolution 1.8A. Northeast Structural Genomics Consortium target NsR236 | | Descriptor: | All0216 protein, POTASSIUM ION | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-10 | | Release date: | 2009-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray structure of All0216 protein from Nostoc sp. PCC 7120 at the resolution 1.8A
To be Published
|
|
4UWF
 
 | | Discovery of (2S)-8-((3R)-3-Methylmorpholin-4-yl)-1-(3-methyl-2-oxo- butyl)-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido(1,2-a)pyrimidin-6- one: a Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors | | Descriptor: | (8S)-9-[3,5-bis(fluoranyl)phenyl]-2-morpholin-4-yl-8-(trifluoromethyl)-7,8-dihydro-6H-pyrimido[1,2-a]pyrimidin-4-one, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT TYPE 3 | | Authors: | Pasquier, B, El-Ahmad, Y, Filoche-Romme, B, Dureuil, C, Fassy, F, Abecassis, P.Y, Mathieu, M, Bertrand, T, Benard, T, Barriere, C, ElBatti, S, Letallec, J.P, Sonnefraud, V, Brollo, M, Delbarre, L, Loyau, V, Pilorge, F, Bertin, L, Richepin, P, Arigon, J, Labrosse, J.R, Clement, J, Durand, F, Combet, R, Perraut, P, Leroy, V, Gay, F, Lefrancois, D, Bretin, F, Marquette, J.P, Michot, N, Caron, A, Castell, C, Schio, L, McCort, G, Goulaouic, H, Garcia-Echeverria, C, Ronan, B. | | Deposit date: | 2014-08-12 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Discovery of (2S)-8-[(3R)-3-Methylmorpholin-4-Yl]-1-(3-Methyl-2-Oxobutyl)-2-(Trifluoromethyl)-3,4-Dihydro-2H-Pyrimido[1,2-A]Pyrimidin-6-One: A Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors.
J.Med.Chem., 58, 2015
|
|
3K6I
 
 | |
3K92
 
 | | Crystal structure of a E93K mutant of the majour Bacillus subtilis glutamate dehydrogenase RocG | | Descriptor: | DI(HYDROXYETHYL)ETHER, NAD-specific glutamate dehydrogenase | | Authors: | Gunka, K, Newman, J.A, Commichau, F.M, Herzberg, C, Rodrigues, C, Hewitt, L, Lewis, R.J, Stulke, J. | | Deposit date: | 2009-10-15 | | Release date: | 2010-06-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional dissection of a trigger enzyme: mutations of the bacillus subtilis glutamate dehydrogenase RocG that affect differentially its catalytic activity and regulatory properties
J.Mol.Biol., 400, 2010
|
|
3K9Z
 
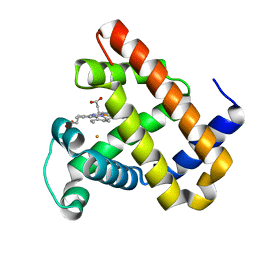 | | Rational Design of a Structural and Functional Nitric Oxide Reductase | | Descriptor: | FE (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yeung, N, Lin, Y.-W, Gao, Y.-G, Zhao, X, Russell, B.S, Lei, L, Miner, K.D, Robinson, H, Lu, Y. | | Deposit date: | 2009-10-16 | | Release date: | 2009-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Rational design of a structural and functional nitric oxide reductase.
Nature, 462, 2009
|
|
3JS3
 
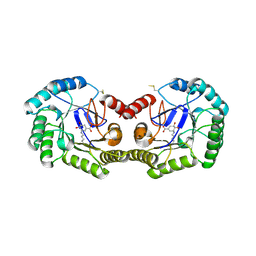 | | Crystal structure of type I 3-dehydroquinate dehydratase (aroD) from Clostridium difficile with covalent reaction intermediate | | Descriptor: | 3-AMINO-4,5-DIHYDROXY-CYCLOHEX-1-ENECARBOXYLATE, 3-dehydroquinate dehydratase | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Dubrovska, I, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-09 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into the mechanism of type I dehydroquinate dehydratases from structures of reaction intermediates.
J.Biol.Chem., 286, 2011
|
|
6FE5
 
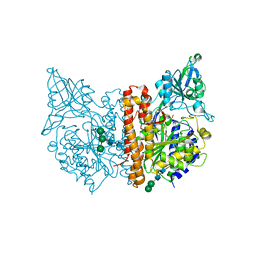 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) - the E424M inactive mutant, in complex with a inhibitor JHU 2249 | | Descriptor: | (2~{S})-2-[[(2~{S})-4-methyl-1-oxidanyl-1-oxidanylidene-pentan-2-yl]carbamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Novakova, Z, Motlova, L. | | Deposit date: | 2017-12-29 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and computational basis for potent inhibition of glutamate carboxypeptidase II by carbamate-based inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
4UWG
 
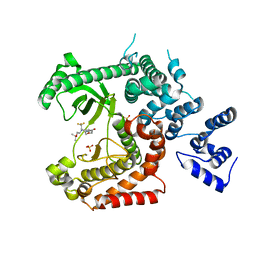 | | Discovery of (2S)-8-((3R)-3-Methylmorpholin-4-yl)-1-(3-methyl-2-oxo- butyl)-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido(1,2-a)pyrimidin-6- one: a Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors | | Descriptor: | (8S)-2-(morpholin-4-yl)-9-[2-(propan-2-yloxy)ethyl]-8-(trifluoromethyl)-6,7,8,9-tetrahydro-4H-pyrimido[1,2-a]pyrimidin-4-one, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT TYPE 3, SULFATE ION | | Authors: | Pasquier, B, El-Ahmad, Y, Filoche-Romme, B, Dureuil, C, Fassy, F, Abecassis, P.Y, Mathieu, M, Bertrand, T, Benard, T, Barriere, C, ElBatti, S, Letallec, J.P, Sonnefraud, V, Brollo, M, Delbarre, L, Loyau, V, Pilorge, F, Bertin, L, Richepin, P, Arigon, J, Labrosse, J.R, Clement, J, Durand, F, Combet, R, Perraut, P, Leroy, V, Gay, F, Lefrancois, D, Bretin, F, Marquette, J.P, Michot, N, Caron, A, Castell, C, Schio, L, McCort, G, Goulaouic, H, Garcia-Echeverria, C, Ronan, B. | | Deposit date: | 2014-08-12 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of (2S)-8-[(3R)-3-Methylmorpholin-4-Yl]-1-(3-Methyl-2-Oxobutyl)-2-(Trifluoromethyl)-3,4-Dihydro-2H-Pyrimido[1,2-A]Pyrimidin-6-One: A Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors.
J.Med.Chem., 58, 2015
|
|
3K2V
 
 | | Structure of the CBS pair of a putative D-arabinose 5-phosphate isomerase from Klebsiella pneumoniae subsp. pneumoniae. | | Descriptor: | CYTIDINE 5'-MONOPHOSPHATE 3-DEOXY-BETA-D-GULO-OCT-2-ULO-PYRANOSONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Cuff, M.E, Volkart, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-30 | | Release date: | 2009-12-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the CBS pair of a putative D-arabinose 5-phosphate isomerase from Klebsiella pneumoniae subsp. pneumoniae.
TO BE PUBLISHED
|
|
6F5L
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a inhibitor JHU2379 | | Descriptor: | (2~{S})-2-[[(2~{S})-4-methyl-1-oxidanyl-1-oxidanylidene-pentan-2-yl]carbamoyloxy]pentanedioic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Novakova, Z, Motlova, L. | | Deposit date: | 2017-12-01 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and computational basis for potent inhibition of glutamate carboxypeptidase II by carbamate-based inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
3K5R
 
 | |
3K5Y
 
 | | Crystal structure of FBF-2/gld-1 FBEa complex | | Descriptor: | 5'-R(*UP*GP*UP*GP*CP*CP*AP*UP*A)-3', Fem-3 mRNA-binding factor 2 | | Authors: | Wang, Y, Opperman, L, Wickens, M, Hall, T.M.T. | | Deposit date: | 2009-10-08 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3K6D
 
 | |
3JT0
 
 | | Crystal Structure of the C-terminal fragment (426-558) Lamin-B1 from Homo sapiens, Northeast Structural Genomics Consortium Target HR5546A | | Descriptor: | Lamin-B1 | | Authors: | Kuzin, A, Abashidze, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-11 | | Release date: | 2009-09-22 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Northeast Structural Genomics Consortium Target HR5546A
To be Published
|
|
8C8T
 
 | | cryo-EM structure of BG505 SOSIP.664 HIV-1 Env trimer in complex with bNAbs EPTC112 and 3BNC117 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Baquero, E, Molinos-Albert, L, Mouquet, H. | | Deposit date: | 2023-01-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Anti-V1/V3-glycan broadly HIV-1 neutralizing antibodies in a post-treatment controller.
Cell Host Microbe, 31, 2023
|
|
3JZI
 
 | | Crystal structure of biotin carboxylase from E. Coli in complex with benzimidazole series | | Descriptor: | 7-amino-2-[(2-chlorobenzyl)amino]-1-{[(1S,2S)-2-hydroxycycloheptyl]methyl}-1H-benzimidazole-5-carboxamide, Biotin carboxylase | | Authors: | Cheng, C, Shipps, G.W, Yang, Z, Sun, B, Kawahata, N, Soucy, K, Soriano, A, Orth, P, Xiao, L, Mann, P, Black, T. | | Deposit date: | 2009-09-23 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery and optimization of antibacterial AccC inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
