2VFN
 
 | | Low Temperature Structure of P22 Tailspike Protein Fragment (109-666), Mutant V125A | | Descriptor: | BIFUNCTIONAL TAIL PROTEIN, CALCIUM ION, GLYCEROL, ... | | Authors: | Becker, M, Mueller, J.J, Heinemann, U, Seckler, R. | | Deposit date: | 2007-11-05 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Side-Chain Stacking and Beta-Helix Stability in P22 Tailspike Protein
To be Published
|
|
2WNH
 
 | | Crystal Structure Analysis of Klebsiella sp ASR1 Phytase | | Descriptor: | 3-PHYTASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Bohm, K, Mueller, J.J, Heinemann, U. | | Deposit date: | 2009-07-09 | | Release date: | 2010-04-28 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Klebsiella Sp. Asr1 Phytase Suggests Substrate Binding to a Preformed Active Site that Meets the Requirements of a Plant Rhizosphere Enzyme.
FEBS J., 277, 2010
|
|
2WLB
 
 | | Adrenodoxin-like ferredoxin Etp1fd(516-618) of Schizosaccharomyces pombe mitochondria | | Descriptor: | ELECTRON TRANSFER PROTEIN 1, MITOCHONDRIAL, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Mueller, J.J, Hannemann, F, Schiffler, B, Bernhardt, R, Heinemann, U. | | Deposit date: | 2009-06-23 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Thermodynamic Characterization of the Adrenodoxin-Like Domain of the Electron-Transfer Protein Etp1 from Schizosaccharomyces Pombe.
J.Inorg.Biochem., 105, 2011
|
|
2WU0
 
 | |
2WNI
 
 | |
2VFM
 
 | | Low Temperature Structure of P22 Tailspike Protein Fragment (109-666) | | Descriptor: | BIFUNCTIONAL TAIL PROTEIN, CALCIUM ION, GLYCEROL, ... | | Authors: | Becker, M, Mueller, J.J, Heinemann, U, Seckler, R. | | Deposit date: | 2007-11-05 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Side-Chain Stacking and Beta-Helix Stability in P22 Tailspike Protein
To be Published
|
|
2W7N
 
 | | Crystal Structure of KorA Bound to Operator DNA: Insight into Repressor Cooperation in RP4 Gene Regulation | | Descriptor: | 5'-D(*AP*AP*TP*GP*TP*TP*TP*AP*GP*CP *TP*AP*AP*AP*CP*AP*AP*G)-3', 5'-D(*CP*BRUP*BRUP*GP*TP*TP*TP*AP*GP*CP*TP*AP *AP*AP*CP*AP*BRUP*T)-3', TRFB TRANSCRIPTIONAL REPRESSOR PROTEIN | | Authors: | Koenig, B, Mueller, J.J, Lanka, E, Heinemann, U. | | Deposit date: | 2008-12-23 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Kora Bound to Operator DNA: Insight Into Repressor Cooperation in Rp4 Gene Regulation
Nucleic Acids Res., 37, 2009
|
|
2WAL
 
 | | Crystal Structure of human GADD45gamma | | Descriptor: | GROWTH ARREST AND DNA-DAMAGE-INDUCIBLE PROTEIN GADD45 GAMMA, MALONIC ACID | | Authors: | Bhattacharya, S, Mueller, J.J, Roske, Y, Turnbull, A.P, Quedenau, C, Goetz, F, Buessow, K, Heinemann, U. | | Deposit date: | 2009-02-09 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Human Gadd45Gamma
To be Published
|
|
1AXK
 
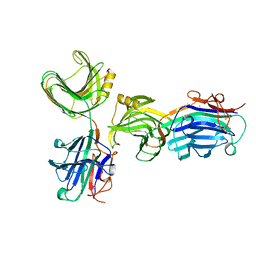 | | ENGINEERED BACILLUS BIFUNCTIONAL ENZYME GLUXYN-1 | | Descriptor: | CALCIUM ION, GLUXYN-1 | | Authors: | Ay, J, Heinemann, U. | | Deposit date: | 1997-10-16 | | Release date: | 1999-05-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of the Bacillus hybrid enzyme GluXyn-1: native-like jellyroll fold preserved after insertion of autonomous globular domain.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1AJK
 
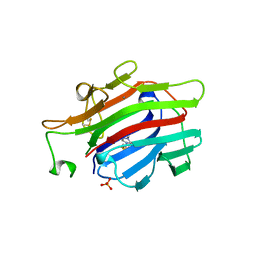 | | CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-84 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, ... | | Authors: | Ay, J, Heinemann, U. | | Deposit date: | 1997-05-06 | | Release date: | 1998-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and properties of de novo circularly permuted 1,3-1,4-beta-glucanases.
Proteins, 30, 1998
|
|
1AYF
 
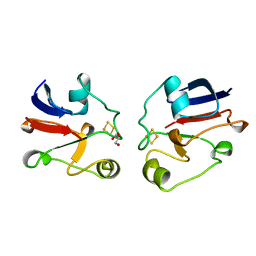 | | BOVINE ADRENODOXIN (OXIDIZED) | | Descriptor: | ADRENODOXIN, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL | | Authors: | Mueller, A, Mueller, J.J, Heinemann, U. | | Deposit date: | 1997-11-03 | | Release date: | 1998-12-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | New aspects of electron transfer revealed by the crystal structure of a truncated bovine adrenodoxin, Adx(4-108).
Structure, 6, 1998
|
|
1BYH
 
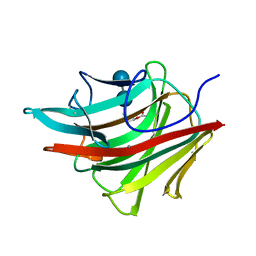 | | MOLECULAR AND ACTIVE-SITE STRUCTURE OF A BACILLUS (1-3,1-4)-BETA-GLUCANASE | | Descriptor: | CALCIUM ION, HYBRID, N-BUTANE, ... | | Authors: | Keitel, T, Heinemann, U. | | Deposit date: | 1992-12-31 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular and active-site structure of a Bacillus 1,3-1,4-beta-glucanase.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1AJO
 
 | | CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-127 | | Descriptor: | CALCIUM ION, CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-127 | | Authors: | Ay, J, Heinemann, U. | | Deposit date: | 1997-05-07 | | Release date: | 1998-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structures and properties of de novo circularly permuted 1,3-1,4-beta-glucanases.
Proteins, 30, 1998
|
|
4BIR
 
 | | RIBONUCLEASE T1: FREE HIS92GLN MUTANT | | Descriptor: | CALCIUM ION, GUANYL-SPECIFIC RIBONUCLEASE T1 | | Authors: | Doumen, J, Steyaert, J. | | Deposit date: | 1998-01-13 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of histidine-40 in ribonuclease T1 catalysis: three-dimensionalstructures of the partially active His40Lys mutant.
Biochemistry, 31, 1992
|
|
5BIR
 
 | |
6XVT
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-PPPPTEDDL-NH2 | | Descriptor: | ACY-SC1-SC2-SC3-SC4-SC5-NME, NITRATE ION, Protein enabled homolog, ... | | Authors: | Barone, M, Le Cong, K, Roske, Y. | | Deposit date: | 2020-01-22 | | Release date: | 2020-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4RNT
 
 | |
3BIR
 
 | |
1RN1
 
 | | THREE-DIMENSIONAL STRUCTURE OF GLN 25-RIBONUCLEASE T1 AT 1.84 ANGSTROMS RESOLUTION: STRUCTURAL VARIATIONS AT THE BASE RECOGNITION AND CATALYTIC SITES | | Descriptor: | RIBONUCLEASE T1 ISOZYME, SULFATE ION | | Authors: | Arni, R.K, Pal, G.P, Ravichandran, K.G, Tulinsky, A, Walz Junior, F.G, Metcalf, P. | | Deposit date: | 1991-11-22 | | Release date: | 1994-01-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Three-dimensional structure of Gln25-ribonuclease T1 at 1.84-A resolution: structural variations at the base recognition and catalytic sites.
Biochemistry, 31, 1992
|
|
8Q1X
 
 | | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | Authors: | Roske, Y, Daumke, O, Damme, M. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation.
Nucleic Acids Res., 52, 2024
|
|
8Q1K
 
 | | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | Authors: | Roske, Y, Daumke, O, Damme, M. | | Deposit date: | 2023-07-31 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation.
Nucleic Acids Res., 52, 2024
|
|
6RCJ
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-15]-OMe | | Descriptor: | GLYCEROL, NITRATE ION, Protein enabled homolog, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2019-04-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6RCF
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-15]-OH | | Descriptor: | 2-[(3~{a}~{R},6~{R},8~{a}~{S})-1-[(3~{S},6~{R},8~{a}~{S})-1'-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8~{a}-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-6-ethyl-8-oxidanylidene-3,3~{a},6,8~{a}-tetrahydro-2~{H}-pyrrolo[2,3-c]azepin-7-yl]ethanoic acid, NITRATE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2019-04-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6RD2
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-1]-TEDEL-NH2 | | Descriptor: | (3~{S},7~{R},10~{R},13~{S})-4-[(3~{S},6~{R},8~{a}~{S})-1'-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8~{a}-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxylic acid, GLYCEROL, NITRATE ION, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2019-04-12 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XXR
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-PPPPTEDEA-NH2 | | Descriptor: | Ac-[2-Cl-F]-PPPPTEDEA-NH2, NITRATE ION, Protein enabled homolog | | Authors: | Barone, M, Le Cong, K, Roske, Y. | | Deposit date: | 2020-01-28 | | Release date: | 2020-11-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
