6IGG
 
 | | Crystal structure of FT condition 1 | | Descriptor: | 1,2-ETHANEDIOL, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
6IGJ
 
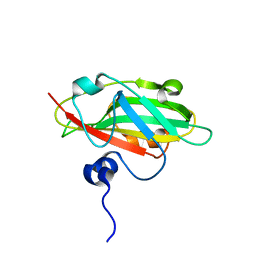 | | Crystal structure of FT condition 4 | | Descriptor: | MAGNESIUM ION, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
1VFF
 
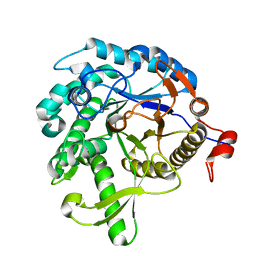 | |
6IGH
 
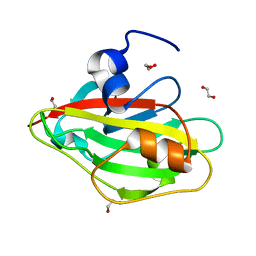 | | Crystal structure of FT condition3 | | Descriptor: | 1,2-ETHANEDIOL, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
6IGI
 
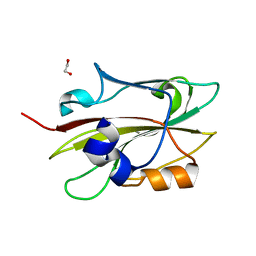 | | Crystal structure of FT condition 2 | | Descriptor: | 1,2-ETHANEDIOL, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
1D7F
 
 | | CRYSTAL STRUCTURE OF ASPARAGINE 233-REPLACED CYCLODEXTRIN GLUCANOTRANSFERASE FROM ALKALOPHILIC BACILLUS SP. 1011 DETERMINED AT 1.9 A RESOLUTION | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLUCANOTRANSFERASE | | Authors: | Ishii, N, Haga, K, Yamane, K, Harata, K. | | Deposit date: | 1999-10-18 | | Release date: | 2000-03-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of asparagine 233-replaced cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011 determined at 1.9 A resolution.
J.Mol.Recog., 13, 2000
|
|
2D42
 
 | | Crystal structure analysis of a non-toxic crystal protein from Bacillus thuringiensis | | Descriptor: | non-toxic crystal protein | | Authors: | Akiba, T, Higuchi, K, Mizuki, E, Ekino, K, Shin, T, Ohba, M, Kanai, R, Harata, K. | | Deposit date: | 2005-10-05 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Nontoxic crystal protein from Bacillus thuringiensis demonstrates a remarkable structural similarity to beta-pore-forming toxins
Proteins, 63, 2006
|
|
3WJJ
 
 | | Crystal structure of IIb selective Fc variant, Fc(P238D), in complex with FcgRIIb | | Descriptor: | Ig gamma-1 chain C region, Low affinity immunoglobulin gamma Fc region receptor II-b, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kadono, S, Mimoto, F, Katada, H, Igawa, T, Kuramochi, T, Muraoka, M, Wada, Y, Haraya, K, Miyazaki, T, Hattori, K. | | Deposit date: | 2013-10-10 | | Release date: | 2013-11-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineered antibody Fc variant with selectively enhanced Fc gamma RIIb binding over both Fc gamma RIIaR131 and Fc gamma RIIaH131.
Protein Eng.Des.Sel., 26, 2013
|
|
3WN6
 
 | | Crystal structure of alpha-amylase AmyI-1 from Oryza sativa | | Descriptor: | Alpha-amylase, CALCIUM ION, D(-)-TARTARIC ACID, ... | | Authors: | Ochiai, A, Sugai, H, Harada, K, Tanaka, S, Ishiyama, Y, Ito, K, Tanaka, T, Uchiumi, T, Taniguchi, M, Mitsui, T. | | Deposit date: | 2013-12-05 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of alpha-amylase from Oryza sativa: molecular insights into enzyme activity and thermostability
Biosci.Biotechnol.Biochem., 78, 2014
|
|
1IC4
 
 | | CRYSTAL STRUCTURE OF HYHEL-10 FV MUTANT(HD32A)-HEN LYSOZYME COMPLEX | | Descriptor: | IGG1 FAB CHAIN H, LYSOZYME BINDING IG KAPPA CHAIN, LYSOZYME C | | Authors: | Shiroishi, M, Yokota, A, Tsumoto, K, Kondo, H, Nishimiya, Y, Horii, K, Matsushima, M, Ogasahara, K, Yutani, K, Kumagai, I. | | Deposit date: | 2001-03-30 | | Release date: | 2001-07-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural evidence for entropic contribution of salt bridge formation to a protein antigen-antibody interaction: the case of hen lysozyme-HyHEL-10 Fv complex.
J.Biol.Chem., 276, 2001
|
|
1IC7
 
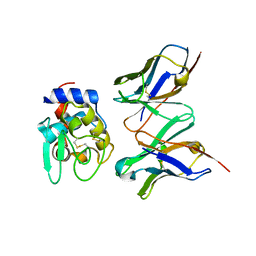 | | CRYSTAL STRUCTURE OF HYHEL-10 FV MUTANT(HD32A99A)-HEN LYSOZYME COMPLEX | | Descriptor: | IGG1 FAB CHAIN H, LYSOZYME BINDING IG KAPPA CHAIN, LYSOZYME C | | Authors: | Shiroishi, M, Yokota, A, Tsumoto, K, Kondo, H, Nishimiya, Y, Horii, K, Matsushima, M, Ogasahara, K, Yutani, K, Kumagai, I. | | Deposit date: | 2001-03-30 | | Release date: | 2001-07-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evidence for entropic contribution of salt bridge formation to a protein antigen-antibody interaction: the case of hen lysozyme-HyHEL-10 Fv complex.
J.Biol.Chem., 276, 2001
|
|
1IC5
 
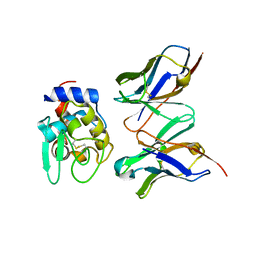 | | CRYSTAL STRUCTURE OF HYHEL-10 FV MUTANT(HD99A)-HEN LYSOZYME COMPLEX | | Descriptor: | IGG1 FAB CHAIN H, LYSOZYME BINDING IG KAPPA CHAIN, LYSOZYME C | | Authors: | Shiroishi, M, Yokota, A, Tsumoto, K, Kondo, H, Nishimiya, Y, Horii, K, Matsushima, M, Ogasahara, K, Yutani, K, Kumagai, I. | | Deposit date: | 2001-03-30 | | Release date: | 2001-07-18 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural evidence for entropic contribution of salt bridge formation to a protein antigen-antibody interaction: the case of hen lysozyme-HyHEL-10 Fv complex.
J.Biol.Chem., 276, 2001
|
|
5AY8
 
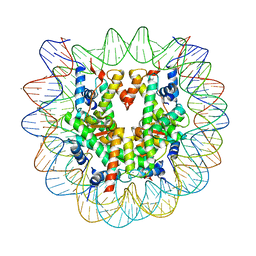 | | Crystal structure of human nucleosome containing H3.Y | | Descriptor: | CHLORIDE ION, DNA (146-MER), H3.Y, ... | | Authors: | Kujirai, T, Horikoshi, N, Sato, K, Maehara, K, Machida, S, Osakabe, A, Kimura, H, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2015-08-10 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and function of human histone H3.Y nucleosome
Nucleic Acids Res., 44, 2016
|
|
1GD9
 
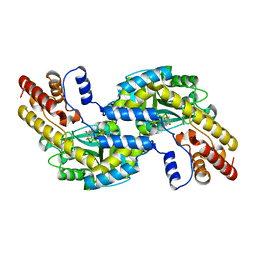 | | CRYSTALL STRUCTURE OF PYROCOCCUS PROTEIN-A1 | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Harata, K, Matsui, I, Kuramitsu, S. | | Deposit date: | 2000-09-22 | | Release date: | 2001-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Temperature dependence of the enzyme-substrate recognition mechanism.
J.Biochem., 129, 2001
|
|
1I75
 
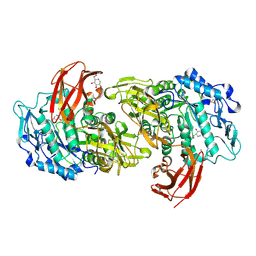 | | CRYSTAL STRUCTURE OF CYCLODEXTRIN GLUCANOTRANSFERASE FROM ALKALOPHILIC BACILLUS SP.#1011 COMPLEXED WITH 1-DEOXYNOJIRIMYCIN | | Descriptor: | 1-DEOXYNOJIRIMYCIN, CALCIUM ION, CYCLODEXTRIN GLUCANOTRANSFERASE | | Authors: | Kanai, R, Haga, K, Yamane, K, Harata, K. | | Deposit date: | 2001-03-08 | | Release date: | 2001-04-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011 complexed with 1-deoxynojirimycin at 2.0 A resolution.
J.Biochem.(Tokyo), 129, 2001
|
|
5CPI
 
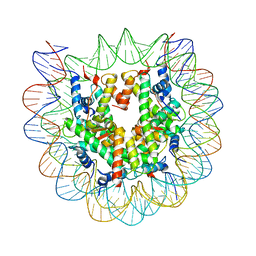 | | Nucleosome containing unmethylated Sat2R DNA | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Osakabe, A, Arimura, Y, Adachi, F, Maehara, K, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Influence of DNA methylation on positioning and DNA flexibility of nucleosomes with pericentric satellite DNA.
Open Biology, 5, 2015
|
|
5CPK
 
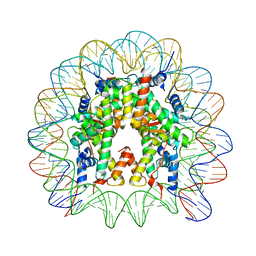 | | Nucleosome containing methylated Sat2L DNA | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Osakabe, A, Arimura, Y, Adachi, F, Maehara, K, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.632 Å) | | Cite: | Influence of DNA methylation on positioning and DNA flexibility of nucleosomes with pericentric satellite DNA.
Open Biology, 5, 2015
|
|
1IZO
 
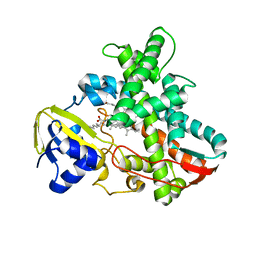 | | Cytochrome P450 BS beta Complexed with Fatty Acid | | Descriptor: | Cytochrome P450 152A1, PALMITOLEIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, D.S, Yamada, A, Sugimoto, H, Matsunaga, I, Ogura, H, Ichihara, K, Adachi, S, Park, S.Y, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-10 | | Release date: | 2003-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Recognition and Molecular Mechanism of Fatty Acid Hydroxylation by Cytochrome P450 from Bacillus subtilis. CRYSTALLOGRAPHIC, SPECTROSCOPIC, AND MUTATIONAL STUDIES.
J.Biol.Chem., 278, 2003
|
|
5CPJ
 
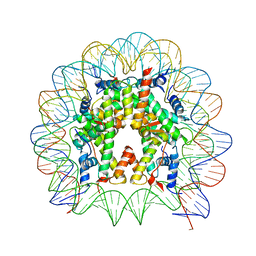 | | Nucleosome containing methylated Sat2R DNA | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Osakabe, A, Arimura, Y, Adachi, F, Maehara, K, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Influence of DNA methylation on positioning and DNA flexibility of nucleosomes with pericentric satellite DNA.
Open Biology, 5, 2015
|
|
1GDE
 
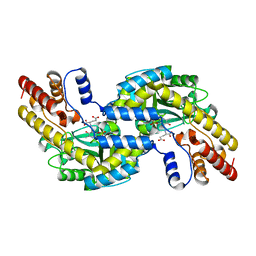 | | CRYSTAL STRUCTURE OF PYROCOCCUS PROTEIN A-1 E-FORM | | Descriptor: | ASPARTATE AMINOTRANSFERASE, GLUTAMIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Harata, K, Matsui, I, Kuramitsu, S. | | Deposit date: | 2000-09-23 | | Release date: | 2001-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Temperature dependence of the enzyme-substrate recognition mechanism.
J.Biochem., 129, 2001
|
|
1ON8
 
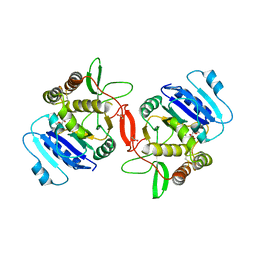 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) with UDP and GlcUAb(1-3)Galb(1-O)-naphthalenelmethanol an acceptor substrate analog | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-27 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1OM2
 
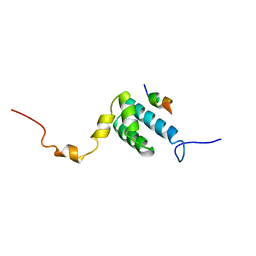 | | SOLUTION NMR STRUCTURE OF THE MITOCHONDRIAL PROTEIN IMPORT RECEPTOR TOM20 FROM RAT IN A COMPLEX WITH A PRESEQUENCE PEPTIDE DERIVED FROM RAT ALDEHYDE DEHYDROGENASE (ALDH) | | Descriptor: | PROTEIN (MITOCHONDRIAL ALDEHYDE DEHYDROGENASE), PROTEIN (MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM20) | | Authors: | Abe, Y, Shodai, T, Muto, T, Mihara, K, Torii, H, Nishikawa, S, Endo, T, Kohda, D. | | Deposit date: | 1999-04-23 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20.
Cell(Cambridge,Mass.), 100, 2000
|
|
1OMZ
 
 | | crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) in complex with UDPGalNAc | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2),
a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1OMX
 
 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2 | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1ON6
 
 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminotransferase (EXTL2) in complex with UDPGlcNAc | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-27 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
