6JNO
 
 | | RXRa structure complexed with CU-6PMN | | Descriptor: | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid, Retinoic acid receptor RXR-alpha | | Authors: | Kawasaki, M, Nakano, S, Motoyama, T, Yamada, S, Watanabe, M, Takamura, Y, Fujihara, M, Tokiwa, H, Kakuta, H, Ito, S. | | Deposit date: | 2019-03-17 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Competitive Binding Assay with an Umbelliferone-Based Fluorescent Rexinoid for Retinoid X Receptor Ligand Screening.
J.Med.Chem., 62, 2019
|
|
9ILB
 
 | | HUMAN INTERLEUKIN-1 BETA | | Descriptor: | PROTEIN (HUMAN INTERLEUKIN-1 BETA) | | Authors: | Yu, B, Blaber, M, Gronenborn, A.M, Clore, G.M, Caspar, D.L.D. | | Deposit date: | 1998-10-22 | | Release date: | 1999-01-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Disordered water within a hydrophobic protein cavity visualized by x-ray crystallography.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
8IDQ
 
 | | Crystal structure of reducing-end xylose-releasing exoxylanase in GH30 from Talaromyces cellulolyticus with xylose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nakamichi, Y, Watanabe, M, Fujii, T, Inoue, H, Morita, T. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of reducing-end xylose-releasing exoxylanase in subfamily 7 of glycoside hydrolase family 30.
Proteins, 91, 2023
|
|
8IDP
 
 | | Crystal structure of reducing-end xylose-releasing exoxylanase in GH30 from Talaromyces cellulolyticus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamichi, Y, Watanabe, M, Fujii, T, Inoue, H, Morita, T. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of reducing-end xylose-releasing exoxylanase in subfamily 7 of glycoside hydrolase family 30.
Proteins, 91, 2023
|
|
8IGU
 
 | |
8IGW
 
 | | Hexameric Ring Complex of Engineered V1-ATPase bound to 4 ADPs: A3(De)3_(ADP)3cat,1non-cat, Hexameric Ring Complex of Engineered V1-ATPase bound to 5 ADPs: A3(De)3_(ADP)3cat,2non-cat | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Kosugi, T, Tanabe, M, Koga, N. | | Deposit date: | 2023-02-21 | | Release date: | 2023-07-12 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Design of allosteric sites into rotary motor V 1 -ATPase by restoring lost function of pseudo-active sites.
Nat.Chem., 15, 2023
|
|
8IGV
 
 | | Hexameric Ring Complex of Engineered V1-ATPase bound to 5 ADPs: A3(De)3_(ADP-Pi)1cat(ADP)2cat,2non-cat | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kosugi, T, Tanabe, M, Koga, N. | | Deposit date: | 2023-02-21 | | Release date: | 2023-07-12 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Design of allosteric sites into rotary motor V 1 -ATPase by restoring lost function of pseudo-active sites.
Nat.Chem., 15, 2023
|
|
2YU0
 
 | | Solution structures of the PAAD_DAPIN domain of mus musculus interferon-activatable protein 205 | | Descriptor: | Interferon-activable protein 205 | | Authors: | Sato, M, Tochio, N, Koshiba, S, Watanabe, M, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the PAAD_DAPIN domain of mus musculus interferon-activatable protein 205
To be Published
|
|
9JIX
 
 | | Crystal structure of de novo designed ATPase, PL2x4_2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, AMMONIUM ION, ... | | Authors: | Kosugi, T, Tanabe, M, Koga, N. | | Deposit date: | 2024-09-12 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | De novo design of ATPase based on a blueprint optimized for harboring the P-loop motif.
Protein Sci., 34, 2025
|
|
3ZIM
 
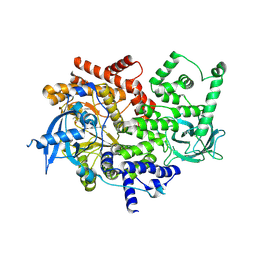 | | Discovery of a potent and isoform-selective targeted covalent inhibitor of the lipid kinase PI3Kalpha | | Descriptor: | 1-[4-[[2-(1H-indazol-4-yl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidin-6-yl]methyl]piperazin-1-yl]-6-methyl-hept-5-ene-1,4- dione, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT ALPHA ISOFORM | | Authors: | Nacht, M, Qiao, L, Sheets, M.P, Martin, T.S, Labenski, M, Mazdiyasni, H, Karp, R, Zhu, Z, Chaturvedi, P, Bhavsar, D, Niu, D, Westlin, W, Petter, R.C, Medikonda, A.P, Jestel, A, Blaesse, M, Singh, J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-02-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of a Potent and Isoform-Selective Targeted Covalent Inhibitor of the Lipid Kinase Pi3Kalpha
J.Med.Chem., 56, 2013
|
|
4AKR
 
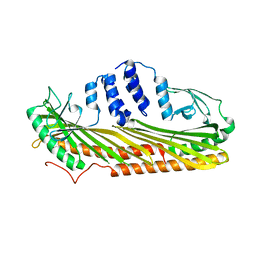 | | Crystal Structure of the cytoplasmic actin capping protein Cap32_34 from Dictyostelium discoideum | | Descriptor: | F-ACTIN-CAPPING PROTEIN SUBUNIT ALPHA, F-ACTIN-CAPPING PROTEIN SUBUNIT BETA | | Authors: | Eckert, C, Goretzki, A, Faberova, M, Kollmar, M. | | Deposit date: | 2012-02-28 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conservation and Divergence between Cytoplasmic and Muscle-Specific Actin Capping Proteins: Insights from the Crystal Structure of Cytoplasmic CAP32/34 from Dictyostelium Discoideum.
Bmc Struct.Biol., 12, 2012
|
|
6JNR
 
 | | RXRa structure complexed with CU-6PMN and SRC1 peptide. | | Descriptor: | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid, HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Retinoic acid receptor RXR-alpha | | Authors: | Kawasaki, M, Nakano, S, Motoyama, T, Yamada, S, Watanabe, M, Takamura, Y, Fujihara, M, Tokiwa, H, Kakuta, H, Ito, S. | | Deposit date: | 2019-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RXRa structure complexed with CU-6PMN and SRC1 peptide.
To Be Published
|
|
1IT8
 
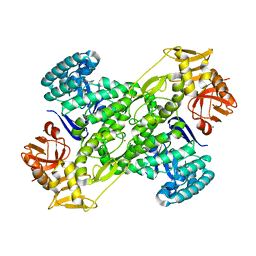 | | Crystal structure of archaeosine tRNA-guanine transglycosylase from Pyrococcus horikoshii complexed with archaeosine precursor, preQ0 | | Descriptor: | 2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDINE-5-CARBONITRILE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ishitani, R, Nureki, O, Fukai, S, Kijimoto, T, Nameki, N, Watanabe, M, Kondo, H, Sekine, M, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-01-11 | | Release date: | 2002-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of archaeosine tRNA-guanine transglycosylase.
J.Mol.Biol., 318, 2002
|
|
1IT7
 
 | | Crystal structure of archaeosine tRNA-guanine transglycosylase complexed with guanine | | Descriptor: | Archaeosine tRNA-guanine transglycosylase, GUANINE, MAGNESIUM ION, ... | | Authors: | Ishitani, R, Nureki, O, Fukai, S, Kijimoto, T, Nameki, N, Watanabe, M, Kondo, H, Sekine, M, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-01-11 | | Release date: | 2002-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of archaeosine tRNA-guanine transglycosylase.
J.Mol.Biol., 318, 2002
|
|
6V9S
 
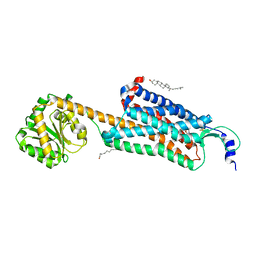 | | Structure-based development of subtype-selective orexin 1 receptor antagonists | | Descriptor: | CHOLESTEROL, OLEIC ACID, Orexin receptor type 1,GlgA glycogen synthase chimera, ... | | Authors: | Hellmann, J, Drabek, M, Yin, J, Huebner, H, Kraus, F, Proell, T, Weikert, D, Kolb, P, Rosenbaum, D.M, Gmeiner, P. | | Deposit date: | 2019-12-16 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-based development of a subtype-selective orexin 1 receptor antagonist.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1IQ8
 
 | | Crystal Structure of archaeosine tRNA-guanine transglycosylase from Pyrococcus horikoshii | | Descriptor: | ARCHAEOSINE TRNA-GUANINE TRANSGLYCOSYLASE, MAGNESIUM ION, ZINC ION | | Authors: | Ishitani, R, Nureki, O, Fukai, S, Kijimoto, T, Nameki, N, Watanabe, M, Kondo, H, Sekine, M, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-07-09 | | Release date: | 2002-05-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of archaeosine tRNA-guanine transglycosylase.
J.Mol.Biol., 318, 2002
|
|
3WQ8
 
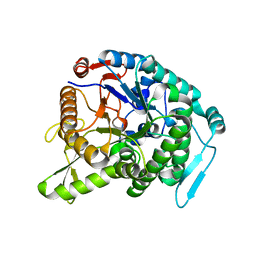 | | Monomer structure of hyperthermophilic beta-glucosidase mutant forming a dodecameric structure in the crystal form | | Descriptor: | Beta-glucosidase | | Authors: | Nakabayashi, M, Kataoka, M, Watanabe, M, Ishikawa, K. | | Deposit date: | 2014-01-23 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Monomer structure of a hyperthermophilic beta-glucosidase mutant forming a dodecameric structure in the crystal form.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
1M9H
 
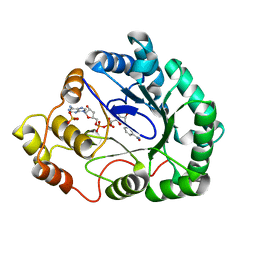 | | Corynebacterium 2,5-DKGR A and Phe 22 replaced with Tyr (F22Y), Lys 232 replaced with Gly (K232G), Arg 238 replaced with His (R238H)and Ala 272 replaced with Gly (A272G)in presence of NADH cofactor | | Descriptor: | 2,5-diketo-D-gluconic acid reductase A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Sanli, G, Blaber, M. | | Deposit date: | 2002-07-29 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural alteration of cofactor specificity in Corynebacterium 2,5-diketo-D-gluconic acid reductase
Protein Sci., 13, 2004
|
|
4M4D
 
 | | Crystal structure of lipopolysaccharide binding protein | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipopolysaccharide-binding protein | | Authors: | Eckert, J.K, Kim, Y.J, Kim, J.I, Gurtler, K, Oh, D.Y, Ploeg, A.H, Pickkers, P, Lundvall, L, Hamann, L, Giamarellos-Bourboulis, E, Kubarenko, A.V, Weber, A.N, Kabesch, M, Kumpf, O, An, H.J, Lee, J.O, Schumann, R.R. | | Deposit date: | 2013-08-07 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.909 Å) | | Cite: | The crystal structure of lipopolysaccharide binding protein reveals the location of a frequent mutation that impairs innate immunity.
Immunity, 39, 2013
|
|
4QKR
 
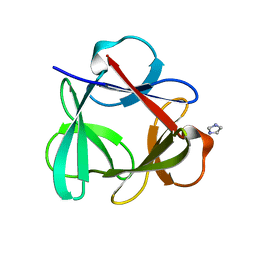 | |
4QKS
 
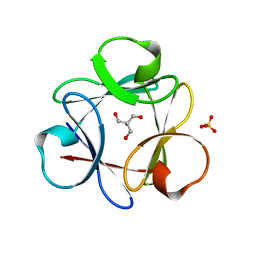 | | Crystal Structure of 6xTrp/PV2: de novo designed beta-trefoil architecture with symmetric primary structure (L22W/L44W/L64W/L85W/L108W/L132W his Primitive Version 2) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DE NOVO PROTEIN 6XTRP/PV2, SULFATE ION | | Authors: | Longo, L.M, Tenorio, C.A, Blaber, M. | | Deposit date: | 2014-06-09 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A single aromatic core mutation converts a designed "primitive" protein from halophile to mesophile folding.
Protein Sci., 24, 2015
|
|
1M16
 
 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Terminal His Tag and Leu 44 Replaced with Phe (L44F), Leu 73 Replaced with Val (L73V), Val 109 Replaced with Leu (V109L) and Cys 117 Replaced with Val (C117V). | | Descriptor: | FORMIC ACID, SULFATE ION, acidic fibroblast growth factor | | Authors: | Brych, S.R, Kim, J, Spielmann, G.L, Logan, T.M, Blaber, M. | | Deposit date: | 2002-06-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Accommodation of a highly symmetric core within a symmetric protein superfold
Protein Sci., 12, 2003
|
|
4Q91
 
 | |
1NZK
 
 | | Crystal Structure of a Multiple Mutant (L44F, L73V, V109L, L111I, C117V) of Human Acidic Fibroblast Growth Factor | | Descriptor: | Acidic Fibroblast Growth Factor, FORMIC ACID, SULFATE ION | | Authors: | Brych, S.R, Kim, J, Logan, T.M, Blaber, M. | | Deposit date: | 2003-02-18 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Accommodation of a highly symmetric core within a symmetric protein superfold.
Protein Sci., 12, 2003
|
|
5XL0
 
 | | met-aquo form of sperm whale myoglobin reconstituted with 7-PF, a heme possesseing CF3 group as side chain | | Descriptor: | Myoglobin, SULFATE ION, fluorinated heme | | Authors: | Kanai, Y, Harada, A, Shibata, T, Nishimura, R, Namiki, K, Watanabe, M, Nakamura, S, Yumoto, F, Senda, T, Suzuki, A, Neya, S, Yamamoto, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Characterization of Heme Orientational Disorder in a Myoglobin Reconstituted with a Trifluoromethyl-Group-Substituted Heme Cofactor
Biochemistry, 56, 2017
|
|
