4K69
 
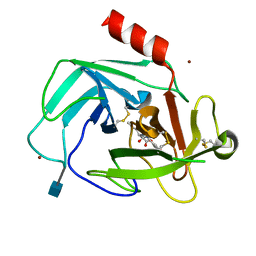 | | Crystal Structure of Human Chymase in Complex with Fragment Linked Benzimidazolone Inhibitor: (3S)-3-{3-[(6-bromo-2-oxo-2,3-dihydro-1H-indol-4-yl)methyl]-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl}hexanoic acid | | Descriptor: | (3S)-3-{3-[(6-bromo-2-oxo-2,3-dihydro-1H-indol-4-yl)methyl]-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl}hexanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chymase, ... | | Authors: | Collins, B.K, Padyana, A.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Potent, Selective Chymase Inhibitors via Fragment Linking Strategies.
J.Med.Chem., 56, 2013
|
|
4KRU
 
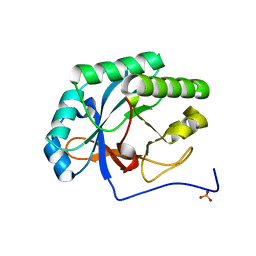 | |
4K60
 
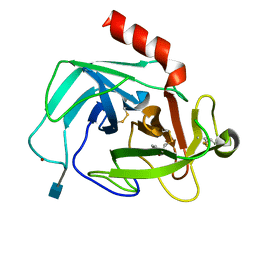 | | Crystal Structure of Human Chymase in Complex with Fragment 6-bromo-1,3-dihydro-2H-indol-2-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-bromo-1,3-dihydro-2H-indol-2-one, Chymase, ... | | Authors: | Collins, B.K, Padyana, A.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Potent, Selective Chymase Inhibitors via Fragment Linking Strategies.
J.Med.Chem., 56, 2013
|
|
4K2Y
 
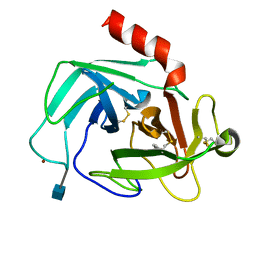 | | Crystal Structure of Human Chymase in Complex with Fragment Inhibitor 6-chloro-1,3-dihydro-2H-indol-2-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-1,3-dihydro-2H-indol-2-one, Chymase, ... | | Authors: | Collins, B.K, Padyana, A.K. | | Deposit date: | 2013-04-09 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Potent, Selective Chymase Inhibitors via Fragment Linking Strategies.
J.Med.Chem., 56, 2013
|
|
4KRT
 
 | |
5W08
 
 | | A/Texas/50/2012(H3N2) Influenza hemagglutinin in complex with K03.12 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | McCarthy, K.R, Harrison, S.C. | | Deposit date: | 2017-05-30 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Memory B Cells that Cross-React with Group 1 and Group 2 Influenza A Viruses Are Abundant in Adult Human Repertoires.
Immunity, 48, 2018
|
|
4OL9
 
 | |
1WDS
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose, ... | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
1WDP
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
4PCJ
 
 | |
1WDR
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose, ... | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
1WDQ
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
3A9H
 
 | | Crystal Structure of PQQ-dependent sugar dehydrogenase holo-form | | Descriptor: | CALCIUM ION, PYRROLOQUINOLINE QUINONE, Putative uncharacterized protein, ... | | Authors: | Sakuraba, H, Yokono, K, Yoneda, K, Ohshima, T. | | Deposit date: | 2009-10-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic properties and crystal structure of quinoprotein aldose sugar dehydrogenase from hyperthermophilic archaeon Pyrobaculum aerophilum
Arch.Biochem.Biophys., 502, 2010
|
|
3A9G
 
 | | Crystal Structure of PQQ-dependent sugar dehydrogenase apo-form | | Descriptor: | CALCIUM ION, Putative uncharacterized protein, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Sakuraba, H, Yokono, K, Yoneda, K, Ohshima, T. | | Deposit date: | 2009-10-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Catalytic properties and crystal structure of quinoprotein aldose sugar dehydrogenase from hyperthermophilic archaeon Pyrobaculum aerophilum
Arch.Biochem.Biophys., 502, 2010
|
|
6RCI
 
 | | Crystal structure of REXO2 | | Descriptor: | Oligoribonuclease, mitochondrial | | Authors: | Spahr, H, Nicholls, T.J, Larsson, N.G, Gustafsson, C.M. | | Deposit date: | 2019-04-11 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dinucleotide Degradation by REXO2 Maintains Promoter Specificity in Mammalian Mitochondria.
Mol.Cell, 76, 2019
|
|
6RCN
 
 | | Crystal structure of REXO2-D199A-dAdA | | Descriptor: | DNA (5'-D(P*AP*A)-3'), MAGNESIUM ION, Oligoribonuclease, ... | | Authors: | Spahr, H, Nicholls, T.J, Larsson, N.G, Gustafsson, C.M. | | Deposit date: | 2019-04-11 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dinucleotide Degradation by REXO2 Maintains Promoter Specificity in Mammalian Mitochondria.
Mol.Cell, 76, 2019
|
|
6RCL
 
 | | Crystal structure of REXO2-D199A-AA | | Descriptor: | MAGNESIUM ION, Oligoribonuclease, mitochondrial, ... | | Authors: | Spahr, H, Nicholls, T.J, Larsson, N.G, Gustafsson, C.M. | | Deposit date: | 2019-04-11 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Dinucleotide Degradation by REXO2 Maintains Promoter Specificity in Mammalian Mitochondria.
Mol.Cell, 76, 2019
|
|
3DPH
 
 | | HIV-1 capsid C-terminal domain mutant (L211S) | | Descriptor: | HIV-1 CAPSID PROTEIN | | Authors: | Igonet, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2008-07-08 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Residues in the HIV-1 Capsid Assembly Inhibitor Binding Site Are Essential for Maintaining the Assembly-competent Quaternary Structure of the Capsid Protein.
J.Biol.Chem., 283, 2008
|
|
3DS3
 
 | |
3DS4
 
 | |
3DS1
 
 | |
3DS0
 
 | |
3DS2
 
 | | HIV-1 capsid C-terminal domain mutant (Y169A) | | Descriptor: | HIV-1 CAPSID PROTEIN | | Authors: | Vaney, M.-C, Igonet, S, Rey, F.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Residues in the HIV-1 Capsid Assembly Inhibitor Binding Site Are Essential for Maintaining the Assembly-competent Quaternary Structure of the Capsid Protein.
J.Biol.Chem., 283, 2008
|
|
3DS5
 
 | | HIV-1 capsid C-terminal domain mutant (N183A) | | Descriptor: | HIV-1 CAPSID PROTEIN | | Authors: | Igonet, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues in the HIV-1 Capsid Assembly Inhibitor Binding Site Are Essential for Maintaining the Assembly-competent Quaternary Structure of the Capsid Protein.
J.Biol.Chem., 283, 2008
|
|
3DTJ
 
 | | HIV-1 capsid C-terminal domain mutant (E187A) | | Descriptor: | HIV-1 capsid protein | | Authors: | Igonet, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2008-07-15 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Residues in the HIV-1 Capsid Assembly Inhibitor Binding Site Are Essential for Maintaining the Assembly-competent Quaternary Structure of the Capsid Protein.
J.Biol.Chem., 283, 2008
|
|
