[wwPDB] Time-stamped Copies of PDB and EMDB Archives
A snapshot of the PDB Core archive (ftp://ftp.wwpdb.org) as of January 1, 2020 has been added to ftp://snapshots.wwpdb.org and ftp://snapshots.pdbj.org. Snapshots have been archived annually since 2005 to provide readily identifiable data sets for research on the PDB archive.
The directory 20200101 includes the 159,140 experimentally-determined structure and experimental data available at that time. Atomic coordinate and related metadata are available in PDBx/mmCIF, PDB, and XML file formats. The date and time stamp of each file indicates the last time the file was modified. The snapshot of PDB Core archive is 575 GB.
A snapshot of the EMDB Core archive (ftp://ftp.ebi.ac.uk/pub/databases/emdb/) as of January 1, 2020 can be found in ftp://ftp.ebi.ac.uk/pub/databases/emdb_vault/20200101/ and ftp://snapshots.pdbj.org/20200101/. The snapshot of EMDB Core archive contains map files and their metadata within XML files for both released and obsoleted entries (10370 and 130, respectively) and is 1.7 TB in size.
[ wwPDB News ]
[wwPDB] Updated Validation Reports for Released PDB Structures
Updated validation reports for all X-ray, NMR, and 3DEM structures released in the PDB archive are now available.
The updates include new percentile statistics reflecting the state of the PDB archive on December 31, 2019 and calculated map coefficients used for validation report generation.
The updated reports are accessible from the following FTP sites:
- ftp://ftp.wwpdb.org/pub/pdb/validation_reports/ (wwPDB)
- ftp://ftp.rcsb.org/pub/pdb/validation_reports/ (RCSB PDB)
- ftp://ftp.ebi.ac.uk/pub/databases/pdb/validation_reports/ (PDBe)
- ftp://ftp.pdbj.org/pub/pdb/validation_reports/ (PDBj)
A snapshot of the previous version on June 10th is archived at RCSB PDB and PDBj.
These updated wwPDB validation reports provide an assessment of structure quality using widely accepted standards and criteria, recommended by community experts serving in Validation Task Forces.
In addition to recently introduced carbohydrate section and 2D Symbol Nomenclature For Glycan (SNFG) images for oligosaccharides from the carbohydrate remediation project, these reports now incorporate visualization of ligand validation and model fit to electron density maps for X-ray ligands. These include 2-dimensional diagrams of ligands, highlighting geometric validation criteria and, for structures determined by crystallography, 3-dimensional views of electron density.
In addition, EM map analysis, and the fit of EM model to its map volume. FSC curves are also included to compare reported and estimated resolution, where either half maps or FSC data was uploaded.
Validation reports are provided to depositors through OneDep - the wwPDB portal for validation, deposition and biocuration of structure data. The wwPDB partners encourage the use of the stand-alone validation server and the web service API at any time prior to data deposition. Depositors are required to review and accept the reports as part of the data submission process. Validation reports will continue to be developed and improved as we receive recommendations from the expert Validation Task Forces (VTF) for X-ray, NMR, EM, and as we collect feedback from depositors and users.
The wwPDB partners strongly encourage journal editors and referees to request reports from authors as part of the manuscript submission and review process, as already required by Nature, eLife, The Journal of Biological Chemistry, the International Union of Crystallography (IUCr) journals, FEBS journals, Journal of Immunology and Angew Chem Int Ed Engl. Reports are date-stamped, display the wwPDB logo, and represent standardized wwPDB validation.
Further information and sample validation reports are available at wwpdb.org.
Your feedback, comments, and questions are welcome at validation@mail.wwpdb.org.
[ wwPDB News ]
[wwPDB] NMR Restraints Validation Now Available Through OneDep
wwPDB validation reports provided in OneDep now include restraints validation for NMR entries to help users identify potential discrepancies in their data.
Recommendations from the wwPDB NMR Validation Task Force (VTF) on how restraints data should be validated have been published (10.1016/j.str.2013.07.021). Since then, wwPDB has been working with the NMR community to develop the NMR Exchange Format (NEF) dictionary (10.1038/nsmb.3041) to enable validation of restraints data.
In March 2020, OneDep was enabled configured to accept NMR experimental data (chemical shifts and restraints) as a single file, either in NMR-STAR or NEF format. Now, NMR restraints data validation has been incorporated into the wwPDB validation package. Validation Reports generated in OneDep have been extended to include NMR restraints analysis when restraints are deposited in either NEF or NMR-STAR format.
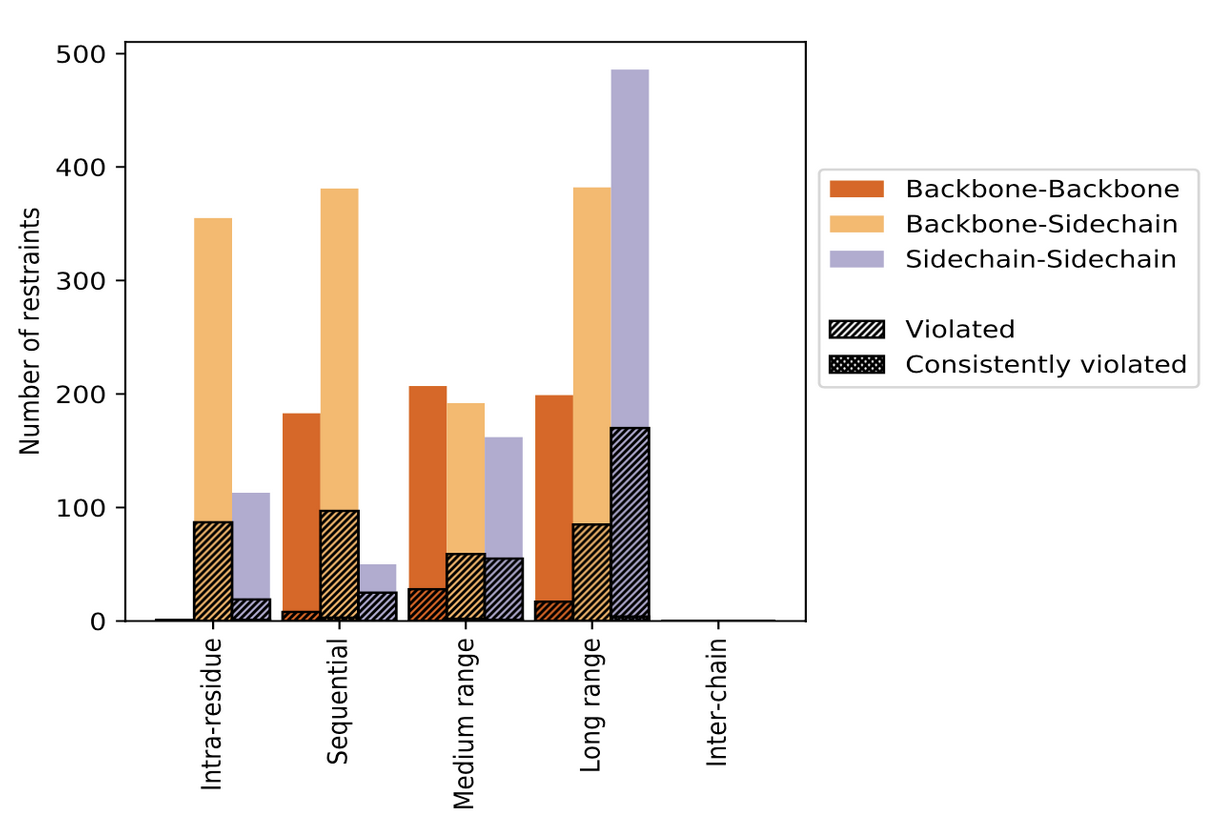
The contents of restraints analysis in the wwPDB validation reports include conformationally restricting restraints, residual restraint violations, average number of distance and dihedral angle restraint violations per model, and distance and dihedral angle violation analysis for each model and ensemble with graphical and tabular statistics.
These features should help both depositors and users to identify potential errors in NMR data.
Additional information about validation reports for NMR entries is available.
If you have any questions or queries about wwPDB validation, then please contact us at validation@mail.wwpdb.org.
 |
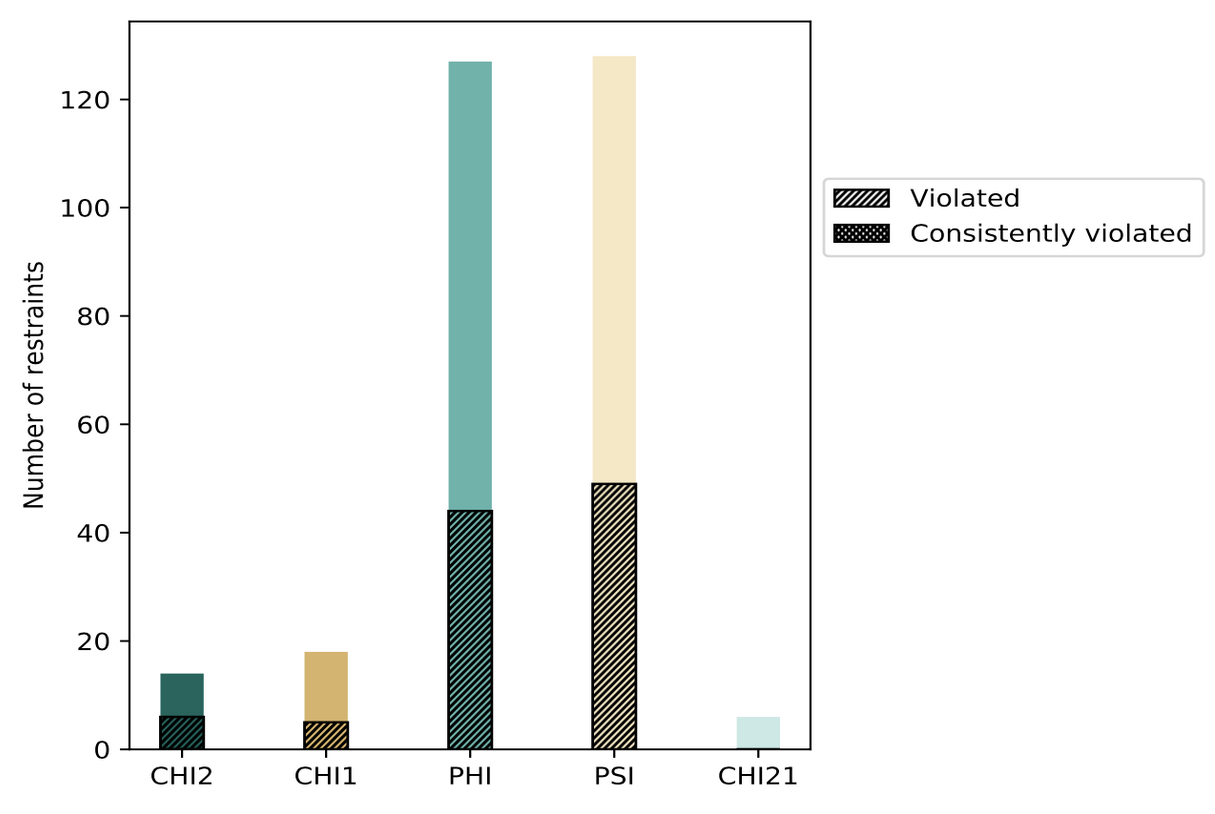 |
| Distribution of distance restraints and violations | Distribution of dihedral-angles and violations |
[ wwPDB News ]
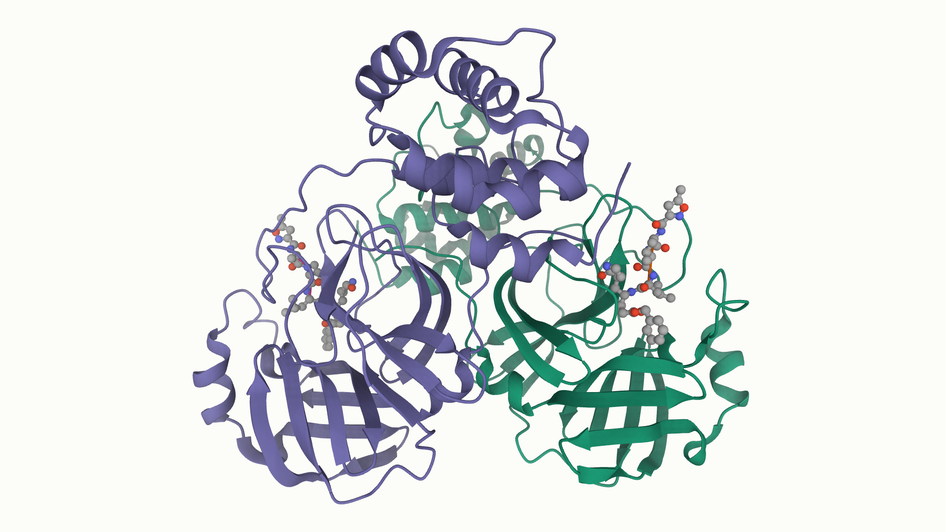
[wwPDB] New Coronavirus Protease Structure Available
PDB data provide a starting point for structure-guided drug discovery
A high-resolution crystal structure of 2019-nCoV coronavirus 3CL hydrolase (Mpro) has been determined by Zihe Rao and Haitao Yang's research team at ShanghaiTech University. Rapid public release of this structure of the main protease of the virus (PDB 6lu7) will enable research on this newly-recognized human pathogen.
Recent emergence of the 2019-nCoV coronavirus has resulted in a WHO-declared public health emergency of international concern. Research efforts around the world are working towards establishing a greater understanding of this particular virus and developing treatments and vaccines to prevent further spread.
While PDB entry 6lu7 is currently the only public-domain 3D structure from this specific coronavirus, the PDB contains structures of the corresponding enzyme from other coronaviruses. The 2003 outbreak of the closely-related Severe Acute Respiratory Syndrome-related coronavirus (SARS) led to the first 3D structures, and today there are more than 200 PDB structures of SARS proteins. Structural information from these related proteins could be vital in furthering our understanding of coronaviruses and in discovery and development of new treatments and vaccines to contain the current outbreak.
The coronavirus 3CL hydrolase (Mpro) enzyme, also known as the main protease, is essential for proteolytic maturation of the virus. It is thought to be a promising target for discovery of small-molecule drugs that would inhibit cleavage of the viral polyprotein and prevent spread of the infection.
Comparison of the protein sequence of the 2019-nCoV coronavirus 3CL hydrolase (Mpro) against the PDB archive identified 95 PDB proteins with at least 90% sequence identity. Furthermore, these related protein structures contain approximately 30 distinct small molecule inhibitors, which could guide discovery of new drugs. Of particular significance for drug discovery is the very high amino acid sequence identity (96%) between the 2019-nCoV coronavirus 3CL hydrolase (Mpro) and the SARS virus main protease (PDB 1q2w). Summary data about these closely-related PDB structures are available (CSV) to help researchers more easily find this information. In addition, the PDB houses 3D structure data for more than 20 unique SARS proteins represented in more than 200 PDB structures, including a second viral protease, the RNA polymerase, the viral spike protein, a viral RNA, and other proteins (CSV).
Public release of the 2019-nCoV coronavirus 3CL hydrolase (Mpro), at a time when this information can prove most vital and valuable, highlights the importance of open and timely availability of scientific data. The wwPDB strives to ensure that 3D biological structure data remain freely accessible for all, while maintaining as comprehensive and accurate an archive as possible. We hope that this new structure, and those from related viruses, will help researchers and clinicians address the 2019-nCoV coronavirus global public health emergency.
[ wwPDB News ]
[wwPDB] 50 Years of the PDB
The PDB was announced on October 20, 1971 in Crystallography: Protein Data Bank Nature New Biology 233: 223 (1971) doi: 10.1038/newbio233223b0.
Fifty years later, the PDB archive contains >170,000 structures of proteins, nucleic acids, and complex assemblies that helps students and researchers understand all aspects of biomedicine and agriculture, from protein synthesis to health and disease. It is managed by the Worldwide PDB (wwPDB) organization that ensures that the PDB is freely and publicly available to the global community.
wwPDB will celebrate this golden anniversary with symposia and events throughout 2021.
Registration for the May 4-5 virtual symposium will be open later this month. This event will include presentations from speakers from around the world who have made tremendous advances in structural biology and bioinformatics. Students and postdoctoral fellows are especially encouraged to attend and will be eligible for poster awards.
During this fiftieth anniversary year, consider supporting the PDB's spirit of openness, cooperation and education with a donation to the wwPDB Foundation.
The wwPDB Foundation was established in 2010 to raise funds in support of the outreach activities of the wwPDB. The Foundation raised funds to help support PDB40, a symposium celebrating the 40th anniversary of the archive; workshops; and educational publications.
[ wwPDB News ]
[wwPDB] Carbohydrate Remediation
An archival-level carbohydrate remediation project that led to the re-release of over 14,000 PDB structures in July 2020. This update includes diverse oligosaccharides: glycosylation; metabolites such as maltose, sucrose, cellulose fragments; glycosaminoglycans, such as fragments of heparin and heparan sulfate; epitope patterns such as A/B blood group antigens and the H-type or Lewis-type stems; and many artificial carbohydrates mimicking or countering natural products (see documentation).
Starting in 2017, this PDB remediation aimed to standardize the biochemical nomenclature of the carbohydrate components following the IUPAC-IUBMB recommendations established by the carbohydrate community (PDF), and to provide uniform representation of oligosaccharides to improve the identification and searchability of oligosaccharides modeled in the PDB structures. During the remediation planning, wwPDB consulted community users and the PDBx/mmCIF Working Group and made data files available on GitHub in early 2020 for community feedback. wwPDB has collaborated with Robert Woods at University of Georgia in US, researchers at The Noguchi Institute and Soka University in Japan, and Thomas Lutteke in Germany to generate uniform linear descriptors for the oligosaccharide sequences.
To achieve these community goals, each oligosaccharide is represented as a branched entity with complete biochemical description and each glycosidic linkage specified. The full representation of carbohydrates is provided in the mmCIF format file, but this is not possible in legacy PDB format files (as the format has been frozen since 2012).
Proper indexing is necessary for branched entity representation and for generation of linear descriptors, hence the ordering (numbering) starts at the reducing end (#1), where the glycosylation occurs, to the non-reducing end in ascending order. Unique chain IDs are assigned to branched entities (oligosaccharides) to avoid residue numbering overlapped with protein residues and to enable consistent numbering for every oligosaccharide. For example, in PDB ID 6WPS, there are 5 oligosaccharides associated with the same protein chain A, the consistent ordering and numbering can only be retained with unique chain ID for each oligosaccharide in both PDBx/mmCIF and PDB format files
For archival consistency, a single-monosaccharide is defined as a non-polymer and treated consistently with other non-polymer ligands in the PDB. A single-monosaccharide occurring at a glycosylation site has a unique chain ID in the PDBx/mmCIF file (_atom_site.label_asym_id) but not in the PDB format file.
Using PDB ID 6WPS as an example, the PDBx/mmCIF data item _atom_site.label_asym_id corresponds to the column #7 in the atom_site coordinates section has an asym ID ‘Y’ for the 1st instance of single-monosaccharide, NAG bound to ASN 61 of protein chain ‘A’. The ‘Y’ value is unique for this monosaccharide. The additional chain ID (_atom_site.auth_asym_id) in the PDBx/mmCIF file that mapped to the PDB format file for this NAG is chain ‘A’, which is consistently represented as any other non-polymer ligands associated with the protein chain A.
#
loop_
_atom_site.group_PDB
_atom_site.id
_atom_site.type_symbol
_atom_site.label_atom_id
_atom_site.label_alt_id
_atom_site.label_comp_id
_atom_site.label_asym_id
_atom_site.label_entity_id
_atom_site.label_seq_id
_atom_site.pdbx_PDB_ins_code
_atom_site.Cartn_x
_atom_site.Cartn_y
_atom_site.Cartn_z
_atom_site.occupancy
_atom_site.B_iso_or_equiv
_atom_site.pdbx_formal_charge
_atom_site.auth_seq_id
_atom_site.auth_comp_id
_atom_site.auth_asym_id
_atom_site.auth_atom_id
_atom_site.pdbx_PDB_model_num
...
HETATM 27655 C C1 . NAG Y 6 . ? 191.103 162.375 206.665 1.00 47.28 ? 1301 NAG A C1 1
HETATM 27656 C C2 . NAG Y 6 . ? 191.067 161.665 208.065 1.00 47.22 ? 1301 NAG A C2 1
HETATM 27657 C C3 . NAG Y 6 . ? 190.138 160.434 207.960 1.00 47.42 ? 1301 NAG A C3 1
HETATM 27658 C C4 . NAG Y 6 . ? 188.730 160.906 207.541 1.00 48.73 ? 1301 NAG A C4 1
HETATM 27659 C C5 . NAG Y 6 . ? 188.838 161.622 206.176 1.00 48.66 ? 1301 NAG A C5 1
HETATM 27660 C C6 . NAG Y 6 . ? 187.494 162.153 205.709 1.00 48.17 ? 1301 NAG A C6 1
HETATM 27661 C C7 . NAG Y 6 . ? 193.233 161.885 209.217 1.00 47.40 ? 1301 NAG A C7 1
HETATM 27662 C C8 . NAG Y 6 . ? 194.594 161.311 209.471 1.00 47.45 ? 1301 NAG A C8 1
HETATM 27663 N N2 . NAG Y 6 . ? 192.418 161.218 208.414 1.00 47.36 ? 1301 NAG A N2 1
HETATM 27664 O O3 . NAG Y 6 . ? 190.069 159.774 209.231 1.00 47.22 ? 1301 NAG A O3 1
HETATM 27665 O O4 . NAG Y 6 . ? 187.867 159.778 207.435 1.00 48.89 ? 1301 NAG A O4 1
HETATM 27666 O O5 . NAG Y 6 . ? 189.760 162.757 206.285 1.00 47.83 ? 1301 NAG A O5 1
HETATM 27667 O O6 . NAG Y 6 . ? 186.953 163.102 206.622 1.00 49.06 ? 1301 NAG A O6 1
HETATM 27668 O O7 . NAG Y 6 . ? 192.879 162.950 209.739 1.00 47.58 ? 1301 NAG A O7 1
...
Author-provided chain ID and residue numbering for oligosaccharides are retained in the PDBx/mmCIF file (_pdbx_branch_scheme.auth_mon_id and _pdbx_branch_scheme.auth_seq_num, respectively). Users can map how carbohydrates are described in the corresponding primary citation to the PDBx/mmCIF files using _pdbx_branch_scheme mapping category. wwPDB strongly encourages depositors to use the wwPDB-assigned chain ID and residue numbers in any publication material.
For example, in PDB entry 6WPS
loop_
_pdbx_branch_scheme.asym_id
_pdbx_branch_scheme.entity_id
_pdbx_branch_scheme.mon_id
_pdbx_branch_scheme.num
_pdbx_branch_scheme.pdb_asym_id
_pdbx_branch_scheme.pdb_mon_id
_pdbx_branch_scheme.pdb_seq_num
_pdbx_branch_scheme.auth_asym_id
_pdbx_branch_scheme.auth_mon_id
_pdbx_branch_scheme.auth_seq_num
_pdbx_branch_scheme.hetero
J 4 NAG 1 I NAG 1 A NAG 1310 n
J 4 NAG 2 I NAG 2 A NAG 1311 n
K 4 NAG 1 J NAG 1 A NAG 1312 n
K 4 NAG 2 J NAG 2 A NAG 1313 n
L 4 NAG 1 K NAG 1 A NAG 1315 n
L 4 NAG 2 K NAG 2 A NAG 1316 n
M 4 NAG 1 M NAG 1 A NAG 1317 n
M 4 NAG 2 M NAG 2 A NAG 1318 n
N 5 NAG 1 N NAG 1 A NAG 1321 n
N 5 NAG 2 N NAG 2 A NAG 1322 n
N 5 BMA 3 N BMA 3 A BMA 1323 n
N 5 MAN 4 N MAN 4 A MAN 1325 n
N 5 MAN 5 N MAN 5 A MAN 1324 n
N 5 FUC 6 N FUC 6 A FUC 1320 n
O 4 NAG 1 O NAG 1 B NAG 1310 n
O 4 NAG 2 O NAG 2 B NAG 1311 n
P 4 NAG 1 P NAG 1 B NAG 1312 n
P 4 NAG 2 P NAG 2 B NAG 1313 n
Q 4 NAG 1 Q NAG 1 B NAG 1315 n
Q 4 NAG 2 Q NAG 2 B NAG 1316 n
R 4 NAG 1 R NAG 1 B NAG 1317 n
R 4 NAG 2 R NAG 2 B NAG 1318 n
S 5 NAG 1 S NAG 1 B NAG 1321 n
S 5 NAG 2 S NAG 2 B NAG 1322 n
S 5 BMA 3 S BMA 3 B BMA 1323 n
S 5 MAN 4 S MAN 4 B MAN 1325 n
S 5 MAN 5 S MAN 5 B MAN 1324 n
S 5 FUC 6 S FUC 6 B FUC 1320 n
...
As some users pointed out, single NAG could be just a part of the glycan that the author chose to build, as most natural N-glycans must have stem of a common core of 5 monosaccharides or its fucosylated version, such as those modeled in the PDB ID 6WPS. However, the PDB is a 3D-atomic coordinate archive in which the model coordinates are built based on supporting experimental data. Therefore, carbohydrates are described as-is in the modeled structures without reference to missing components of the presumed oligosaccharide sequence. If the author only builds a monosaccharide, then this monosaccharide is described as a non-polymer ligand.
Glycosylation annotation has been provided to facilitate searches of all glycosylation sites. A total of 45,000 glycosylation sites have been annotated in _struct_conn.pdbx_role in over 7500 PDB structures to identify all glycosylation sites and the monosaccharides bound at such sites. The annotation specifies the glycosylation sites, the monosaccharide identity and chain IDs in either PDB format or mmCIF format. In PDB ID 6WPS, a user can search N-Glycosylation in ‘_struct_conn.pdbx_role’ and find 16 glycosylation sites between ASN and NAG at chain A alone.
In addition, a total of 1040 carbohydrate ligands were reviewed and their nomenclature has been standardized in the PDB to follow the 1996 IUPAC recommendations. The updates of monosaccharides in the Chemical Component Dictionary include chemical names, synonyms, atom labels, modification vs common sugars tags, chemical types (e.g., isomers), structure feature types (e.g., anomers), and symbol identifiers (e.g., IUPAC condensed symbols). The chemical names provided in PDBx/mmCIF data item _chem_comp.name have been updated uniformly to include stereo- and ring- specific systematic names as described in the IUPAC recommendations. Trivial or common names are annotated in the new PDBx/mmCIF category, _pdbx_chem_comp_synonyms, with one name per row. Both the IUPAC extended-form symbol as described in the section 2-Carb-38.3 of the 1996 recommendation and the condensed-form symbol in 2-Carb-38.4 are provided in the identifier of an PDBx/mmCIF data item _pdbx_chem_comp_identifier.identifier. Carbohydrate features such as isomer, ring size, anomer, and aldose/ketose classification as described in the PDBx/mmCIF category _pdbx_chem_comp_feature.
The wwPDB encourages the community to use PDB/mmCIF format files rather than the frozen legacy PDB file format. The legacy format cannot support large structures. Currently, PDB format-files are not available for large structures that have either more than 62 chains or 99,999 atoms. In addition, the legacy format cannot support ligand ID codes beyond 3-characters, which will be needed in the coming years.
The wwPDB is committed to improving data representation in the PDB archive. Please do not hesitate to contact us at info@wwpdb.org.
[ wwPDB News ]
The number of PDB entries processed by PDBj has reached 40,000
PDBj started processing PDB entries from July 2000, and the total number of PDB entries processed by PDBj since then has reached 40,000 in November 2020(Deposition statistics at wwPDB site). Since the global number of PDB entries processed jointly by RCSB, PDBe and PDBj is about 170,000, about 24% of the PDB entries have been processed by PDBj. We greatly appreciate the continous financial support provided by JST during our 20 year existence.
Please see Newsletter(Vol.21 No.2) for the 20th anniversary of PDBj.
[wwPDB] PDB Turns 49
The PDB was announced on October 20, 1971 in Crystallography: Protein Data Bank Nature New Biology 233: 223 (1971) doi: 10.1038/newbio233223b0.
Today, the PDB archive contains ~170,000 structures of proteins, nucleic acids, and complex assemblies that helps students and researchers understand all aspects of biomedicine and agriculture, from protein synthesis to health and disease. It is managed by the Worldwide PDB (wwPDB) organization that ensures that the PDB is freely and publicly available to the global community.
wwPDB is celebrating by looking ahead to golden anniversary symposia and events planned for 2021. The website wwpdb.org/pdb50 will be updated regularly.
As the fiftieth anniversary approaches, consider supporting the PDB's spirit of openness, cooperation and education with a donation to the wwPDB Foundation.
The wwPDB Foundation was established in 2010 to raise funds in support of the outreach activities of the wwPDB. The Foundation raised funds to help support PDB40, a symposium celebrating the 40th anniversary of the archive; workshops; and educational publications.
The Foundation is chartered as a 501(c)(3) entity exclusively for scientific, literary, charitable, and educational purposes.
[ wwPDB News ]
Explore wwPDB-standardized electron density maps via PDBj EDMap
PDBj now only provides the wwPDB calculated electron density maps through our EDMap service, where we previously used our own generated map data.
From 2020-09-23, all legacy ED Map entries have been replaced by the wwPDB-standardized ED maps calculated from the map coefficients, which were calculated using the Buster software (Global Phasing Ltd), as part of the wwPDB validation pipeline.
The updated map files are available from our FTP site (ftp://ftp.pdbj.org/pub/pdb/validation_reports/) along with the validation reports. In addition, users can also download them from our web interface via PDBj Mine and the PDBj EDmap service.
[wwPDB] An updated PDB-Dev website
An updated PDB-Dev website
A new and improved PDB-Dev website is now available. PDB-Dev is a prototype system which helps wwPDB partners to understand the requirements in archiving integrative structures. It has been built based on recommendations from the wwPDB Integrative/Hybrid Methods (I/HM) Task Force. PDB-Dev currently consists of 37 integrative structures of macromolecular complexes.
The updated PDB-Dev web interface provides dynamic and responsive web pages and includes two new features:
- A new search service that facilitates the retrieval of structures archived in PDB-Dev. The current implementation supports search by macromolecular names, entry identifiers, experimental data types, author names, citations, software and several other keywords. Complex queries with Boolean operators and wildcards are supported. Search results can be downloaded as csv or Excel files.
- Individual entry pages for released entries, highlighting key details and download links for each structure.
The PDB-Dev team welcomes feedback from users. Please send comments and suggestions to pdb-dev@mail.wwpdb.org.
[ wwPDB News ]
[wwPDB] Access Improved Carbohydrate Data at the PDB
A new data representation for carbohydrates in PDB entries and reference data improves the Findability and Interoperability of these molecules in macromolecular structures. The PDB archive now reflects:
- Standardized Chemical Component Dictionary nomenclature following IUPAC-IUBMB recommendations
- Uniform representation for oligosaccharides
- Adoption of glycoscience-community commonly used linear descriptors using community tools
- Annotated glycosylation sites in PDB structures
Detailed information about this project, including a list of remediated PDB entries, is available at the wwPDB website. Developers of software packages that produce, access, or visualize PDB data are encouraged to review this information and adapt their software as soon as possible, as originally highlighted in the February 2020 announcement.
The wwPDB has created a new ‘branched’ entity representation for polysaccharides, describing all the individual monosaccharide components of these in the PDB entry. As part of this process, we have standardized atom nomenclature of >1,000 monosaccharides in the Chemical Component Dictionary (CCD) and applied a branched entity representation to oligosaccharides (>8,000 PDB entries). To guarantee unambiguous chemical description of oligosaccharides in the affected PDB entries, an explicit description of covalent linkage information between their monosaccharide units is included. In addition, wwPDB validation reports provide consistent representation for these oligosaccharides and include 2D representations based on the Symbol Nomenclature for Glycans (SNFG).
To support the remediation of carbohydrate representation, software tools providing linear descriptors were developed in collaboration with the glycoscience community to enable easy translation of PDB data to other representations commonly used by glycobiologists. These include Condense IUPAC from GMML at University of Georgia, WURCS from PDB2Glycan at The Noguchi Institute, Japan, and LINUCS from pdb-care at Germany.
wwPDB has also used this opportunity to improve the organization of chemical synonyms in the CCD by introducing a new _pdbx_chem_comp_synonyms data category. This will enable more comprehensive capture of alternative names for small molecules in the PDB. To minimize disruption to users, the legacy data item, _chem_comp.pdbx_synonyms, will be retained for a transition period through 2021.
The carbohydrate remediation project is a wwPDB collaborative project carried out principally by RCSB PDB at Rutgers, The State University of New Jersey and is funded by NIGMS grant U01 CA221216 in collaboration with Complex Carbohydrate Research Center at the University of Georgia.
If you have any comments or queries regarding these changes, please visit the wwPDB carbohydrate remediation website or contact us at deposit-help@mail.wwpdb.org.
[ wwPDB News ]
We will remove the data files of the previous version of the PDBj Mine RDB from our FTP site
In addition, the dump files and the delta files for the validation report (vrpt) schemas have been made available on our FTP site (ftp://ftp.pdbj.org/mine2/).
2020 July is the 20th anniversary of PDBj.

Protein Data Bank Japan (PDBj) is celebrating the 20th anniversary in July, 2020. It was July 6th, 2000 when PDBj locally processed the 1st deposition to Osaka University, as a part of the JST-BIRD project. While in 2000, only 157 entries were processed locally by PDBj, we grew very rapidly and were able to process 376 entries in 2001 due to the kind technical and scientific help from RCSB PDB, and processed 602 entries in 2002. With the formation of the wwPDB in 2003, Asia and Oceania were assigned to PDBj, but due to a dramatic increase of depositions from Asia, especially main land China, PDBj’s regional area was reassigned to be Asia and the Middle-East in 2016. Since PDBj was established, we have also been building secondary databases such as eF-Site and ProMode, introducing new formats such as PDBMLplus and PDB/RDF, and developed original tools such as Molmil or MagRO. We would like to express our gratitude for the continuous contribution and help from depositors, users and the seamless financial support by JST and from 2017 partly by AMED. PDBj will remain as the central data-hub of Structural Biology in Asia and the Middle-East. We would highly appreciate it, if our community would continue to support and encourage us with our endeavors.
[wwPDB] Coming July 29: Improved Carbohydrate Data at the PDB
PDB data will incorporate a new data representation for carbohydrates in PDB entries and reference data that improves the Findability and Interoperability of these molecules in macromolecular structures. In order to remediate and improve the representation of carbohydrates across the archive, the wwPDB has:
- standardized Chemical Component Dictionary nomenclature following IUPAC-IUBMB recommendations
- provided uniform representation for oligosaccharides
- adopted Glycoscience-community commonly used linear descriptors using community tools
- annotated glycosylation sites in PDB structures
Starting July 29, 2020, users will be able to access the improved data via FTP or wwPDB partner websites. Developers of software packages that produce, access, or visualize PDB data are encouraged to review this information and adapt their software as soon as possible, as originally highlighted in the February 2020 announcement. Detailed information about this project is available at the wwPDB website; lists of impacted entries and chemical components will be published on this page after data release.
The wwPDB has created a new ‘branched’ entity representation for polysaccharides, describing all the individual monosaccharide components of these in the PDB entry. As part of this process, we have standardized atom nomenclature of >1,000 monosaccharides in the Chemical Component Dictionary (CCD) and applied a branched entity representation to oligosaccharides for >8000 PDB entries. To guarantee unambiguous chemical description of oligosaccharides in the affected PDB entries, an explicit description of covalent linkage information between their monosaccharide units is included. In addition, wwPDB validation reports provide consistent representation for these oligosaccharides and include 2D representations based on the Symbol Nomenclature for Glycans (SNFG).
To support the remediation of carbohydrate representation, software tools providing linear descriptors were developed in collaboration with the glycoscience community to enable easy translation of PDB data to other representations commonly used by glycobiologists. These include Condense IUPAC from GMML at University of Georgia, WURCS from PDB2Glycan at The Noguchi Institute, Japan, and LINUCS from pdb-care at Germany.
Furthermore, to ensure continued Findability of 118 common oligosaccharides (e.g., sucrose, Lewis Y antigen), we have expanded the Biologically Interesting molecule Reference Dictionary (BIRD) that contains the covalent linkage information and common synonyms for such molecules.
wwPDB has also used this opportunity to improve the organization of chemical synonyms in the CCD by introducing a new _pdbx_chem_comp_synonyms data category. This will enable more comprehensive capture of alternative names for small molecules in the PDB. To minimize disruption to users, the legacy data item, _chem_comp.pdbx_synonyms, will be retained for a transition period through 2021.
The carbohydrate remediation project is a wwPDB collaborative project that is carried out principally by RCSB PDB at Rutgers, The State University of New Jersey and is funded by NIGMS grant U01 CA221216 in collaboration with Complex Carbohydrate Research Center at the University of Georgia.
If you have any comments or queries regarding these changes, please visit the wwPDB carbohydrate remediation website or contact us at deposit-help@mail.wwpdb.org.
[ wwPDB News ]
We are waiting for donation to the PDBj donation window in the Osaka University Foundation for the Future
We established a PDBj dedicated donation window in the Osaka University Foundation for the Future. We are going to utilize the donated fund to enhance and improve the public relations activities that cannot be covered by research funds from the national and public institutions.
Donation is available from here. Thank you for your cooperation.
Prize of the minister of MEXT in the science and technology field in 2020 was awarded
In 2000, we started the activity of Protein Data Bank Japan (PDBj) at Institute for Protein Research of Osaka University, and have managed the international Protein Data Bank collaborated with Europe and America. On 7th April 2020, the prize of the minister of MEXT (ministry of education, culture, sports, science, and technology) in the science and technology field (promotion part) in 2020 was awarded for Haruki Nakamura (Prof. Emer., IPR, Osaka Univ., the ex-head of PDBj) and Genji Kurisu (Prof., IPR, Osaka Univ., the current head of PDBj), as the work "Contributing for constraction and promoting for integrative usage of the international Protein Data Bank".
Reference: The award winners of the minister of MEXT (ministry of education, culture, sports, science, and technology) prize in the science and technology field in 2020 were decided (Japanese only)
[wwPDB] Streamlining of EMDB’s Map Entry Release Process
Beginning April 15th 2020, the Electron Microscopy Data Bank (EMDB) will end its practice of releasing map entry metadata (so-called XML header files) prior to release of the primary map and associated files.
For new EMDB map depositions, this metadata will not be available publicly in the EMDB FTP public archive until the map is released. For on-hold entries that already have metadata released there will be no change to the available data until the map is released. EMDB strongly encourages depositors to release their map entries as soon as possible.
Abolishing the pre-release of EMDB header files is part of the process to harmonize the policies and procedures of EMDB and wwPDB. For more information about EMDB policies please refer to https://www.ebi.ac.uk/pdbe/emdb/policies.html.
[ wwPDB News ]
COVID-19 featured content has been relased.
PDBj has released the featured page containing all entries directly related to the Novel Coronavirus disease (COVID-19) which is a serious threat to people all over the world.
【COVID-19 featured content】
https://pdbj.org/featured/covid-19
【Molecule of the Month (242):Coronavirus Proteases】
https://pdb101.rcsb.org/motm/242
[wwPDB] Improving carbohydrates in the PDB for 2020
In July 2020, the wwPDB will roll out updated PDB structures and reference data files with standardized representation of carbohydrate molecules, improving the Findability and Interoperability of PDB data. Detailed information about this work is available from the wwPDB website, including PDBx/mmCIF dictionary extensions and over 500 example files. We encourage developers of software packages that produce, access, or visualize PDB data to review this information and adapt their software.
Through collaboration with the glycoscience community, software tools were developed to standardize atom nomenclature of nearly 800 monosaccharides in the Chemical Component Dictionary (CCD) and applied branched polymeric representation to oligo- and polysaccharides within the PDB archive, enabling easy translation to other representations commonly used by glycobiologists. To guarantee unambiguous chemical description of oligo-/polysaccharides in each of the nearly 12,000 affected PDB entries, we have included an explicit description of covalent linkage information between their monomeric units. To ensure continued Findability of common oligosaccharides (e.g., sucrose, Lewis X factor), we have expanded the Biologically Interesting molecule Reference Dictionary (BIRD) which will contain the covalent linkage information and common synonyms for such molecules.
wwPDB is also taking this opportunity to improve the organization of chemical synonyms in the CCD by introducing a new _pdbx_chem_comp_synonyms data category. This will enable more comprehensive capture of alternative names for small molecules in the PDB. To minimize disruption to users, there will be an initial transition period, where the legacy data item, _chem_comp.pdbx_synonyms, will be retained.
.png) PDB structure shown: 1b5f
PDB structure shown: 1b5f
[ wwPDB News ]
[wwPDB] Improve Your Previously-Released Coordinates AND Keep Your Original PDB ID: Phase II
We are pleased to announce that from February 18, 2020 authors (PIs) of released PDB structures can update the model coordinates while retaining the same PDB accession code, thereby preserving the link with the original publication. In this second and final phase of the project we have extended the versioning functionality to structures deposited prior to the roll out of OneDep--the common wwPDB system for deposition, validation, and biocuration.
For entries deposited via OneDep, depositors should log into the corresponding session at deposit.wwpdb.org and submit the request via the OneDep communication panel. For entries deposited via legacy systems, requests should be initiated by sending an email to deposit-help@mail.wwpdb.org and including the PDB code in the subject and body of the email. Once submitted, the revised model will be processed by wwPDB biocurators and the updated version released immediately upon depositor’s approval. Versioning of PDB entries will be limited to changes in the coordinate files, with no changes permitted to the deposited experimental data. To limit the impact on the wwPDB biocuration resources, PDB versioning is currently restricted to one replacement per PDB entry per year, and three entries per Principal Investigator per year. We will review this restriction in 2021.
The most recent version of the entry will be made available in the PDB archive FTP (ftp.wwpdb.org). All major versions of a PDB structure will be retained in the versioned FTP archive (ftp-versioned.wwpdb.org)--more information can be found on the wwPDB website. The structure of the versioned FTP archive has been built allowing for future extension of the PDB code format. PDB entry 1abc would therefore be found in the folder pdb_00001abc.
Changes made to entries during versioning are considered to be either major or minor. Updates to atomic coordinates, polymer sequence, or chemical description trigger a major version increment, while changes to any other categories are classified as minor. Changes introduced are recorded in the PDBx/mmCIF audit categories.
If you have any further queries regarding the process of PDB versioning, please contact the wwPDB at deposit-help@mail.wwpdb.org.
[ wwPDB News ]
Launch of our renewed EDMap service
For new entries, both or either of the wwPDB 2fo-fc and fo-fc maps can be chosen, while for the legacy entries only the original map (PDBj generated 2fo-fc) is available. Furthermore, for download, the structure factor file, the mmCIF formatted map files and our generated MTZ and CCP4 files of the full maps are available for the new wwPDB maps, while for the legacy entries only the structure factor and the generated CCP4 files are available, with the refinement file generated by our previous pipeline being discontinued. Like previously, generated local maps in CCP4 format are available for download for both types.
Regarding the data stored on the FTP, as long as not all entries have been updated with the new wwPDB maps, our legacy data will remain available at ftp://ftp.pdbj.org/edmap/. For the wwPDB data, the maps are available at:
- /pub/pdb/validation_reports/*/*/*_validation_fo-fc_map_coef.cif.gz
- /pub/pdb/validation_reports/*/*/*_validation_2fo-fc_map_coef.cif.gz
While the MTZ and CCP4 files are available at:
- /edmap2/mtz/*_validation_fo-fc_map_coef.mtz
- /edmap2/ccp4/*_validation_fo-fc_map_coef.ccp4
If you have questions or comments regarding our renewed EDmap service, please feel free to contact us.
[wwPDB] Distribution of NMR data in a unified format at the PDB archive
The wwPDB partners are pleased to announce that as of March 2020 the OneDep system will begin accepting upload of NMR experimental data as a single file, either in NMR-STAR or NEF format. This will start the transition from the current practice where distinct types of NMR data such as assigned chemical shifts, restraints, and peak lists are uploaded separately.
NMR-STAR is the official wwPDB format for storing NMR data, supported by an extensive dictionary [GitHub; Ulrich, E. L. et al. (2019) NMR-STAR: comprehensive ontology for representing, archiving and exchanging data from nuclear magnetic resonance spectroscopic experiments Journal of Biomolecular NMR, 73: 5–9. doi: 10.1007/s10858-018-0220-3], while NEF (NMR exchange format; Gutmanas et al. (2015) NMR Exchange Format: a unified and open standard for representation of NMR restraint data Nature Structural & Molecular Biology 22: 433–434 doi: 10.1038/nsmb.3041) is a light-weight format and dictionary, supported by the leading software in NMR structure determination. The use of these two interconvertible standard formats as single data files will simplify the process of deposition, as well as the storage and distribution of this data.
For newly deposited entries accompanied by such a unified data file, the NMR data will be distributed in the PDB FTP area as single files in the NMR-STAR format. A best effort conversion to the NEF format will also be provided. These unified NMR data files will be added to a new FTP directory, “nmr_data” in parallel to the existing directories, nmr_restraints and nmr_chemical shifts. In addition, to support existing users these unified files that contain both restraints and chemical shift data will be copied to the existing directories “nmr_restraints” and “nmr_chemical_shifts”.
A standardized naming convention for NMR unified data will also be developed to simplify access of the relevant NMR data. File naming will start with PDB accession code, followed by nmr_data with format type extension, for example ‘2lcb_nmr_data.nef’ or ‘2lcb_nmr_data.str’.
We plan to begin accepting and distributing NMR data as unified files from March 2020.
[ wwPDB News ]
[wwPDB] EMDB policy and procedures document now available
A comprehensive policy and procedures document for the EMDB archive has been drawn up by the EMDB team in order to ensure consistent and coherent rules for its data. The document is now available to view on the EMDB website.
Since its foundation in 2002, the Electron Microscopy Data Bank (EMDB; https://emdb-empiar.org/) archives publicly available three-dimensional (3D) electron cryo-microscopy (cryo-EM) maps and tomograms of biomacromolecules, their complexes and cellular structures. Following the release of the first eight EMDB entries in 2002, the EMDB archive grew steadily and currently stands at almost 10000 released maps. From 2016, EMDB entries are deposited and processed through the wwPDB OneDep system while the biocuration workload is shared geographically by the EMDB, PDBe, RCSB PDB and PDBj teams.
The policy outlines the requirements for data deposition, accepted formats, entry modifications and release. For example, the policy recommends for single-particle depositions to include a primary map (as shown in the accompanying publication), a raw map (unmasked, unfiltered, unsharpened) and unmasked half-maps, as well as any auxiliary files such as Fourier Shell Correlation (FSC) data. In the document EMDB also advises that the official wwPDB validation report generated after biocuration, which now includes EM map/tomogram validation and, if applicable, map-model validation, is provided to journal editors and referees as part of the manuscript submission and review process.
In response to the cryo-EM community’s increasing demand to make all data publicly available, EMDB strongly encourages the deposition of all atomic models to the Protein Data Bank (PDB), all 3D EM reconstructions to EMDB and all raw data (including tilt series for tomograms) to the Electron Microscopy Public Image Archive (EMPIAR ). Related entries in these archives reference one another, making the deposited data easily discoverable and accessible to the community.
The EMDB team and its umbrella organisation wwPDB, welcome feedback from EMDB users and depositors on the policies and procedures through emdbhelp@ebi.ac.uk.
[ wwPDB News ]