301 new PDB entries have been released on 2026-02-04.
301 new PDB entries have been released on 2026-02-04. 248636 entries are now available in total.
Please refer to PDB / EMDB latest information for other updates.
[wwPDB] EMDB Surpasses 10,000 Entries in a Year
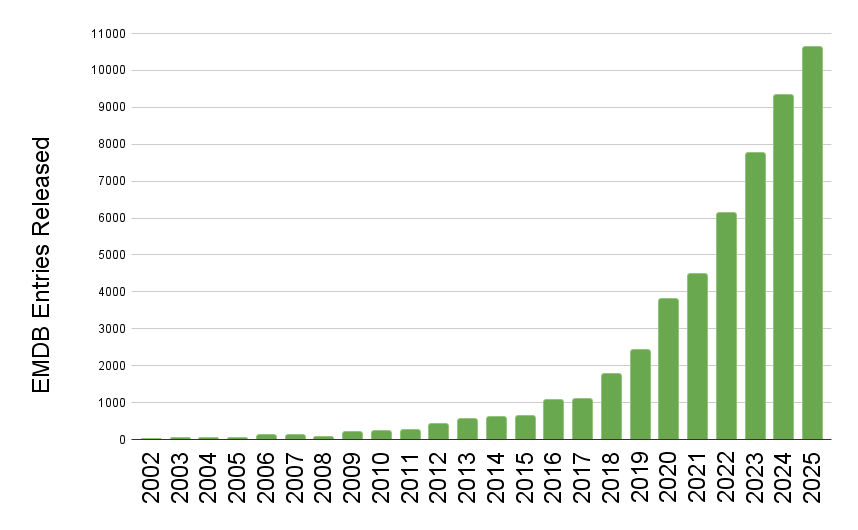
EMDB released entry growth by year
The wwPDB has reached a historic milestone, releasing 10,000 EMDB entries in a single year.
The rate of release of 3DEM data to the public continues at an extraordinary pace, for the first time the wwPDB has seen the release of 10,000 entries from the EMDB in just a single year. To put this in context, in 2019 the EMDB stood at 10,000 entries in total, after 17 years of growth. Now in 2025 due to the current volume of data release, the archive has surpassed 50,000 total entries.
This rapid growth is a testament to the hard work of the community which the wwPDB endeavors to support. It also highlights the ongoing challenge the community is facing with ever more data created requiring streamlined deposition, processing, curation, peer-review, and sustainable & scalable archiving practices. We are excited to continue to be part of structural biology history.
[ wwPDB News ]
[wwPDB] wwPDB Shared Responsibilities on Data Processing
Depositions are automatically distributed to wwPDB deposition centers geographically.
Data processing responsibilities are shared among wwPDB partner sites:
- RCSB PDB is responsible for PDB and EMDB data coming from the Americas
- PDBe and EMDB are responsible for PDB and EMDB data from Europe (except Belarus and Russian Federation) and Africa
- PDBj is responsible for PDB and EMDB data from Asia regions (except China), Belarus, Russian Federation and Oceania
- PDBc is responsible for PDB and EMDB data from China
- BMRB is responsible for BMRB data from the Americas, Africa, and Europe
- BMRBj is responsible for BMRB data from Asia
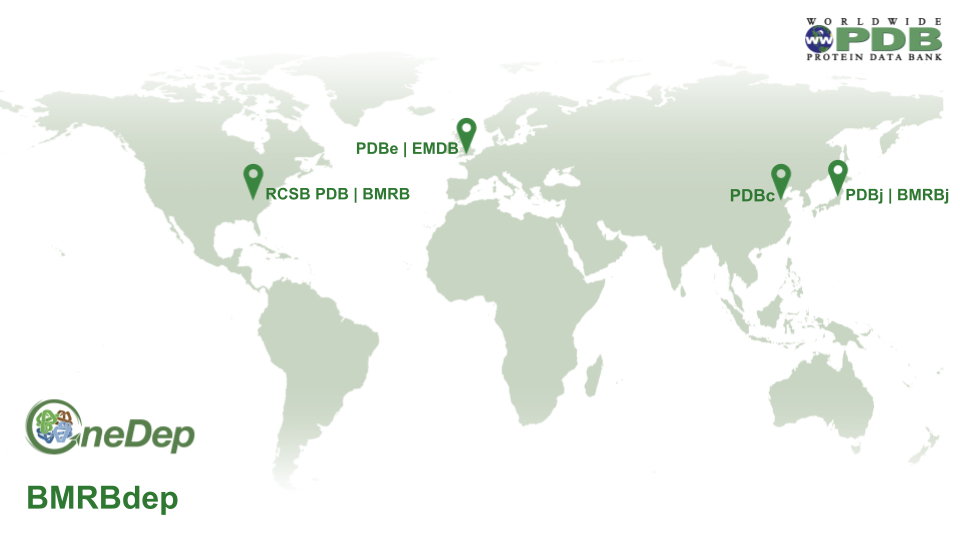 Data processing distribution map
Data processing distribution map[ wwPDB News ]
[wwPDB] Time-stamped Copies of PDB and EMDB Archives
A snapshot of the PDB Core archive (https://files.wwpdb.org, https://s3.rcsb.org) as of January 01, 2026 has been added to https://s3snapshots.rcsb.org (AWS), snapshots.rcsb.org (rsync -rlpy -a -v --delete snapshots.rcsb.org:: .), and ftp://snapshots.pdbj.org. Snapshots have been archived annually since 2005 to provide readily identifiable data sets for research on the PDB archive.
The directory 20260101 includes the 246,905 experimentally-determined structure and experimental data available at that time. Atomic coordinate and related metadata are available in PDBx/mmCIF, PDB, and XML file formats. The date and time stamp of each file indicates the last time the file was modified. The snapshot of PDB Core Archive is 1,583 GB.
A snapshot of the EMDB Core archive (ftp://ftp.ebi.ac.uk/pub/databases/emdb/) as of January 01, 2026 can be found in https://ftp.ebi.ac.uk/pub/databases/emdb_vault/20260101/ and ftp://snapshots.pdbj.org/20260101/. The snapshot includes 52,943 3DEM entries. The Snapshot of the EMDB archive is 28.2 TB in size.
[ wwPDB News ]







