10AD
 
 | | Cryo-EM structure of the human BK channel bound to the agonist NS1619 | | Descriptor: | 3-[2-oxidanyl-5-(trifluoromethyl)phenyl]-6-(trifluoromethyl)-1~{H}-benzimidazol-2-one, CALCIUM ION, Isoform 5 of Calcium-activated potassium channel subunit alpha-1, ... | | Authors: | Gonzalez-Sanabria, N, Contreras, G.F, Perozo, E, Latorre, R. | | Deposit date: | 2026-01-08 | | Release date: | 2026-02-04 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | The BK channel-NS1619 agonist complex reveals molecular insights into allosteric activation gating
Proc.Natl.Acad.Sci.USA, 123, 2026
|
|
10ID
 
 | | Membrane-bound, reversed VP5* trimer (rotavirus spike protein) | | Descriptor: | Outer capsid protein VP4 | | Authors: | de Sautu, M, Leistner, C, Kirchhausen, T, Jenni, S, Harrison, S.C. | | Deposit date: | 2026-01-21 | | Release date: | 2026-02-04 | | Method: | ELECTRON MICROSCOPY (9.54 Å) | | Cite: | Mechanism of membrane perforation in rotavirus cell entry
To Be Published
|
|
10JU
 
 | |
10KE
 
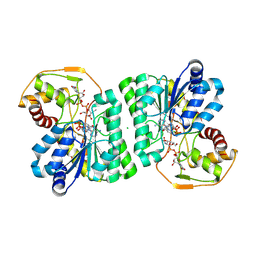 | |
10LG
 
 | |
21FB
 
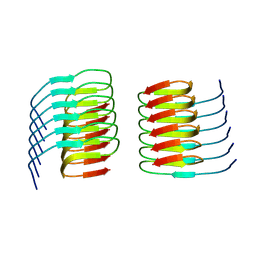 | |
21IE
 
 | |
21IK
 
 | |
21IP
 
 | |
22GA
 
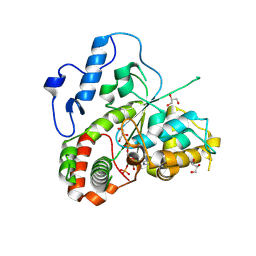 | |
22IY
 
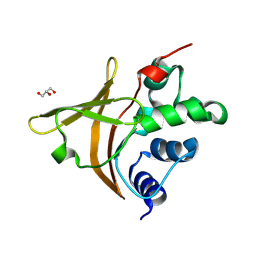 | | Crystal structure of the unliganded microalgal AstaP-pink1 carotenoprotein | | Descriptor: | Astaxanthin-binding pink protein of fasciclin family, GLYCEROL | | Authors: | Sluchanko, N.N, Slonimskiy, Y.B, Varfolomeeva, L.A, Popov, V.O, Boyko, K.M. | | Deposit date: | 2026-01-13 | | Release date: | 2026-02-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the unliganded microalgal AstaP-pink1 carotenoprotein
To Be Published
|
|
22JY
 
 | | P301L/S320F human tau filaments from mouse brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Yanagisawa, H, Kano, M, Kimura, T, Kikkawa, M, Tomita, T. | | Deposit date: | 2026-01-14 | | Release date: | 2026-02-04 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM structure of pro-aggregant P301L/S320F double-mutant tau filaments formed in mouse brains following peripheral AAV delivery
Biorxiv, 2026
|
|
22PS
 
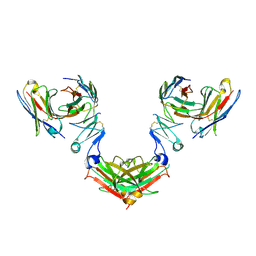 | |
8Z4I
 
 | | crystal structure of Cdn1 in complex with cA3 | | Descriptor: | CALCIUM ION, CARF-domain cantaining dual-activated nuclease, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, W. | | Deposit date: | 2024-04-17 | | Release date: | 2026-02-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural plasticity enables broad cAn binding and dual activation of CRISPR-associated ribonuclease Cdn1.
Nucleic Acids Res., 54, 2026
|
|
9CWY
 
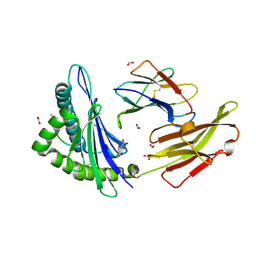 | |
9CWZ
 
 | |
9CX0
 
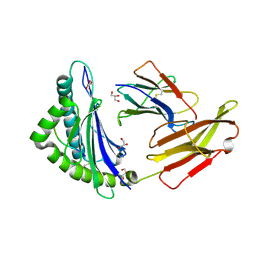 | |
9CX1
 
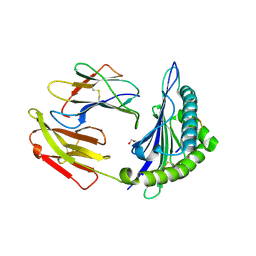 | |
9CX2
 
 | |
9D72
 
 | | CryoEM structure of anti-MHC-I Fab M1/42 complex with H2-Dd | | Descriptor: | Beta-2-microglobulin, Fab M1/42 Heavy Chain, Fab M1/42 Light Chain, ... | | Authors: | Jiang, J, Natarajan, K, Margulies, D.H, Lei, H, Huang, R. | | Deposit date: | 2024-08-16 | | Release date: | 2026-02-04 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | CryoEM structure of anti-MHC-I Fab M1/42 complex with H2-Dd
To Be Published
|
|
9D73
 
 | | CryoEM structure of anti-MHC-I mAb B1.23.2 complex with HLA-B44:05 | | Descriptor: | B1.23.2 Light Chain: VL and C kappa domains, B1.23.2 mAb Heavy Chain: Full-length, Beta-2-microglobulin, ... | | Authors: | Jiang, J, Natarajan, K, Lei, H, Huang, R, Margulies, D.H. | | Deposit date: | 2024-08-16 | | Release date: | 2026-02-04 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | CryoEM structure of anti-MHC-I B1.23.2 complex with HLA-B44:05
To Be Published
|
|
9DMD
 
 | | YscN Yersinia ATPase delta 1-92, R359A | | Descriptor: | GLYCEROL, SULFATE ION, Type 3 secretion system ATPase | | Authors: | Ellis, P.K, Barker, S.A, Dickenson, N.E, Johnson, S.J. | | Deposit date: | 2024-09-13 | | Release date: | 2026-02-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biophysical Characterization of the Yersinia Type Three Secretion System ATPase YscN.
Proteins, 2026
|
|
9E58
 
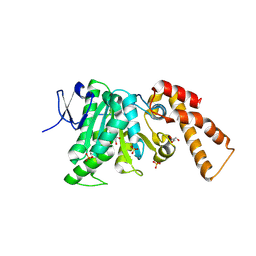 | | YscN Yersinia ATPase delta 1-92 | | Descriptor: | GLYCEROL, SULFATE ION, Type 3 secretion system ATPase | | Authors: | Ellis, P.K, Barker, S.A, Dickenson, N.E, Johnson, S.J. | | Deposit date: | 2024-10-28 | | Release date: | 2026-02-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biophysical Characterization of the Yersinia Type Three Secretion System ATPase YscN.
Proteins, 2026
|
|
9EDR
 
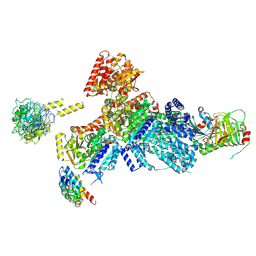 | | Tubulin Cofactors D,E,G,C and Tubulin complex -- TBCC N Terminus Bound to Tubulin | | Descriptor: | Chromosome instability protein 1, GTP-binding protein CIN4, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Taheri, A, Al-bassam, J. | | Deposit date: | 2024-11-17 | | Release date: | 2026-02-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the tubulin cofactors reveal the molecular basis of alpha/beta-tubulin biogenesis.
Nat Commun, 2025
|
|
9EDS
 
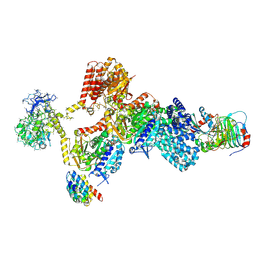 | | Tubulin cofactors D,E,G,C bound to tubulin dimer -- TBCC N terminus unbound | | Descriptor: | Chromosome instability protein 1, GTP-binding protein CIN4, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Taheri, A, Al-bassam, J. | | Deposit date: | 2024-11-18 | | Release date: | 2026-02-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the tubulin cofactors reveal the molecular basis of alpha/beta-tubulin biogenesis.
Nat Commun, 2025
|
|
