9JD2
 
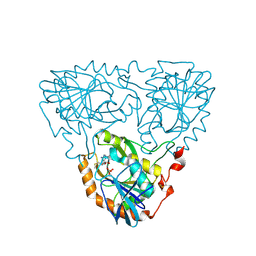 | |
9DDY
 
 | | The Crystal Structure of Geranyltranstransferase from Streptococcus pneumoniae TIGR4 | | Descriptor: | GLYCEROL, Geranyltranstransferase | | Authors: | Kim, Y, Tan, A, Endres, M, Tan, K, Joachimik, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-08-28 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The Crystal Structure of Geranyltranstransferase from Streptococcus pneumoniae TIGR4
To Be Published
|
|
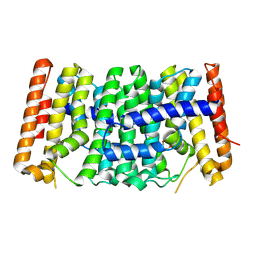
9DCG
 
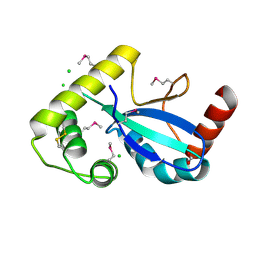 | | Crystal Structure of the Thiol:Disulfide Interchange Protein DsbC from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Shatsman, S, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-08-26 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of the Thiol:Disulfide Interchange Protein DsbC from Vibrio cholerae
To Be Published
|
|
9JA1
 
 | | The RNA polymerase II elongation complex from Saccharomyces cerevisiae | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*GP*CP*TP*CP*CP*TP*TP*CP*TP*CP*CP*CP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*T)-3'), DNA (5'-D(P*TP*GP*GP*GP*AP*GP*AP*AP*GP*GP*AP*GP*C)-3'), ... | | Authors: | Yi, G, Ma, J, Zhang, P. | | Deposit date: | 2024-08-23 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | The RNA polymerase II elongation complex from Saccharomyces cerevisiae
To Be Published
|
|
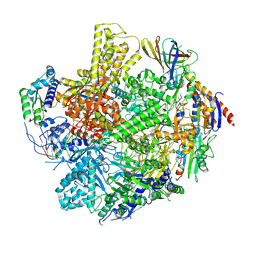
9GJ2
 
 | | Crystal structure of human cathepsin S produced in insect cells in complex with ketoamide 13b | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Cathepsin S, ... | | Authors: | Falke, S, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Reinke, P.Y.A, Guenther, S, Turk, D, Meents, A. | | Deposit date: | 2024-08-20 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of human cathepsin S produced in insect cells in complex with ketoamide 13b
To Be Published
|
|
9J7T
 
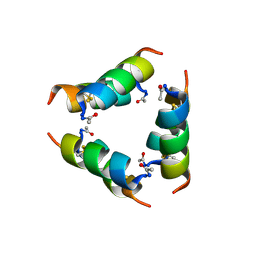 | |
9GGQ
 
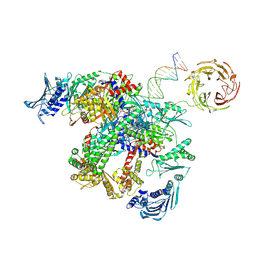 | | E.coli gyrase holocomplex with cleaved chirally wrapped 217 bp DNA fragment and moxifloxacin | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Ghilarov, D, Heddle, J.G, Pabis, M. | | Deposit date: | 2024-08-13 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of chiral wrap and T-segment capture by Escherichia coli DNA gyrase
Proceedings of the National Academy of Sciences USA, 2024
|
|
9D5D
 
 | |
9GG6
 
 | | P301T type II tau filaments from human brain | | Descriptor: | Isoform Tau-F of Microtubule-associated protein tau | | Authors: | Schweighauser, M, Shi, Y, Murzin, A.G, Garringer, H.J, Vidal, R, Murrell, J.R, Erro, M.E, Seelaar, H, Ferrer, I, van Swieten, J.C, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2024-08-13 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Novel tau filament folds in individuals with MAPT mutations P301L and P301T.
Biorxiv, 2024
|
|
9J5G
 
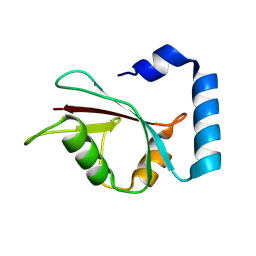 | |
9GG1
 
 | | P301T type I tau filaments from human brain | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Schweighauser, M, Shi, Y, Murzin, A.G, Garringer, H.J, Vidal, R, Murrell, J.R, Erro, M.E, Seelaar, H, Ferrer, I, van Swieten, J.C, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2024-08-12 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Novel tau filament folds in individuals with MAPT mutations P301L and P301T.
Biorxiv, 2024
|
|
9GG0
 
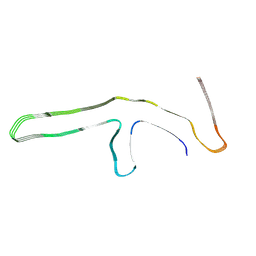 | | P301L tau filaments from human brain | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Schweighauser, M, Shi, Y, Murzin, A.G, Garringer, H.J, Vidal, R, Murrell, J.R, Erro, M.E, Seelaar, H, Ferrer, I, van Swieten, J.C, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2024-08-12 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Novel tau filament folds in individuals with MAPT mutations P301L and P301T.
Biorxiv, 2024
|
|
9D3G
 
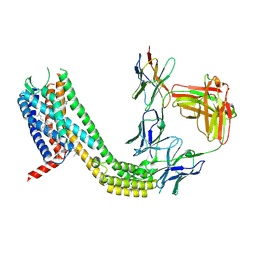 | | Cryo-EM structure of CCR6 bound by SQA1 and OXM1 | | Descriptor: | 1-(4-chlorophenyl)-N-{[(2R)-4-(2,3-dihydro-1H-inden-2-yl)-5-oxomorpholin-2-yl]methyl}cyclopropane-1-carboxamide, 4-[[3,4-bis(oxidanylidene)-2-[[(1~{R})-1-(4-propan-2-ylfuran-2-yl)propyl]amino]cyclobuten-1-yl]amino]-~{N},~{N}-dimethyl-3-oxidanyl-pyridine-2-carboxamide, CCR6, ... | | Authors: | Wasilko, D.J, Wu, H. | | Deposit date: | 2024-08-10 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for CCR6 modulation by allosteric antagonists.
Nat Commun, 15, 2024
|
|
9J4K
 
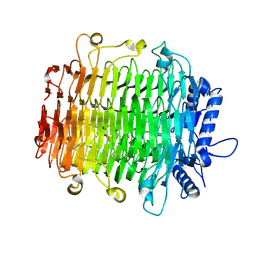 | | Crystal structure of GH9l Inulinfructotransferases (IFTase) in complex with GF2 | | Descriptor: | DFA-III-forming inulin fructotransferase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | Deposit date: | 2024-08-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
9J4J
 
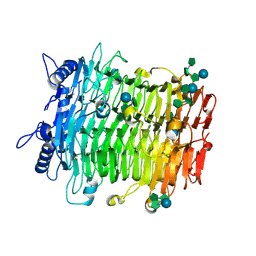 | | Crystal structure of GH9l Inulin fructotransferases(IFTase)incomplex with nystose(F3) | | Descriptor: | DFA-III-forming inulin fructotransferase, beta-D-fructofuranose, beta-D-fructofuranose-(1-1)-beta-D-fructofuranose, ... | | Authors: | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | Deposit date: | 2024-08-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
9J4L
 
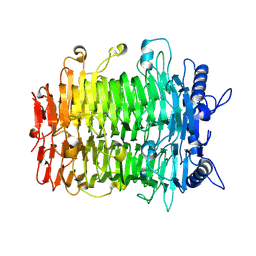 | | Crystal structure of GH9l Inulin fructotransferases (IFTase) | | Descriptor: | DFA-III-forming inulin fructotransferase | | Authors: | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | Deposit date: | 2024-08-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
9D3E
 
 | | Cryo-EM structure of CCR6 bound by SQA1 and OXM2 | | Descriptor: | 4-[[3,4-bis(oxidanylidene)-2-[[(1~{R})-1-(4-propan-2-ylfuran-2-yl)propyl]amino]cyclobuten-1-yl]amino]-~{N},~{N}-dimethyl-3-oxidanyl-pyridine-2-carboxamide, CHOLESTEROL, Human CCR6, ... | | Authors: | Wasilko, D.J, Wu, H. | | Deposit date: | 2024-08-09 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis for CCR6 modulation by allosteric antagonists.
Nat Commun, 15, 2024
|
|
9J4I
 
 | | Crystal structure of GH9l Inulin fructotransferases (IFTase) in compex with fruetosyl nystose (GF4) | | Descriptor: | DFA-III-forming inulin fructotransferase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-[alpha-D-glucopyranose-(1-2)]beta-D-fructofuranose, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-[alpha-D-glucopyranose-(1-2)]beta-D-fructofuranose | | Authors: | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | Deposit date: | 2024-08-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
9D2C
 
 | |
9GEA
 
 | |
9CYP
 
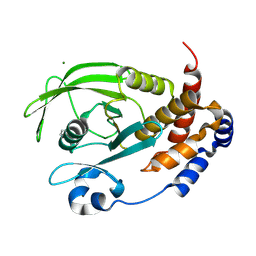 | | Crystal structure of I19V mutant human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
9CYM
 
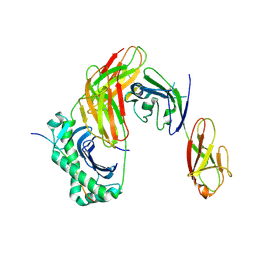 | | Structure of LAG3 bound to the MHC class II molecule I-A(b) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Class-II-associated invariant chain peptide, H-2 class II histocompatibility antigen, ... | | Authors: | Ming, Q, Antfolk, D, Tran, T.H, Luca, V.C. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Structural basis for LAG3 interactions with MHC class II
To Be Published
|
|
9CYO
 
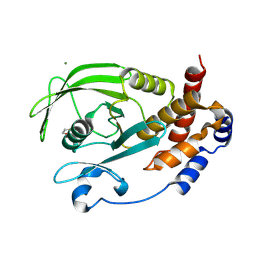 | | Crystal structure of wild-type human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
9CYQ
 
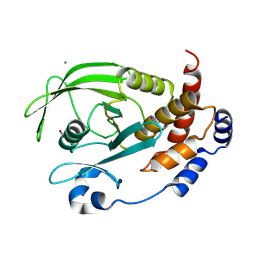 | | Crystal structure of Q78R mutant human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
9CYR
 
 | | Crystal structure of D245G mutant human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
