8PXV
 
 | | Targeting extended blood antigens by Akkermansia muciniphila enzymes unveils a missing link for generating universal donor blood | | Descriptor: | Beta-N-acetylhexosaminidase, GLYCEROL, SODIUM ION, ... | | Authors: | Weikum, J, Jensen, M, Abou Hachem, M, Morth, J.P. | | Deposit date: | 2023-07-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Akkermansia muciniphila exoglycosidases target extended blood group antigens to generate ABO-universal blood.
Nat Microbiol, 9, 2024
|
|
8PXU
 
 | |
8PXT
 
 | | Targeting extended blood antigens by Akkermansia muciniphila enzymes unveils a missing link for generating universal donor blood | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Jensen, M, Abou Hachem, M, Morth, J.P. | | Deposit date: | 2023-07-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Akkermansia muciniphila exoglycosidases target extended blood group antigens to generate ABO-universal blood.
Nat Microbiol, 9, 2024
|
|
8HVB
 
 | |
8HVC
 
 | |
8HVD
 
 | |
8BDP
 
 | |
8BBL
 
 | | SGL a GH20 family sulfoglycosidase | | Descriptor: | Beta-N-acetylhexosaminidase | | Authors: | Dong, M.D, Roth, C.R, Jin, Y.J. | | Deposit date: | 2022-10-13 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 2022
|
|
7WDT
 
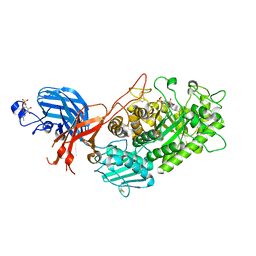 | | 6-sulfo-beta-D-N-acetylglucosaminidase from Bifidobacterium bifidum in complex with GlcNAc-6S | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, ... | | Authors: | Yamada, C, Kashima, T, Fushinobu, S, Katoh, T, Katayama, T. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A bacterial sulfoglycosidase highlights mucin O-glycan breakdown in the gut ecosystem.
Nat.Chem.Biol., 19, 2023
|
|
7WDU
 
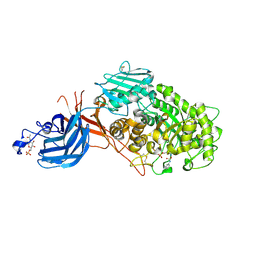 | | 6-sulfo-beta-D-N-acetylglucosaminidase from Bifidobacterium bifidum in complex with PUGNAc-6S | | Descriptor: | Beta-N-acetylhexosaminidase, CALCIUM ION, [[(3R,4R,5S,6R)-3-acetamido-4,5-bis(oxidanyl)-6-(sulfooxymethyl)oxan-2-ylidene]amino] N-phenylcarbamate | | Authors: | Kashima, T, Yamada, C, Fushinobu, S, Katoh, T, Katayama, T. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | A bacterial sulfoglycosidase highlights mucin O-glycan breakdown in the gut ecosystem.
Nat.Chem.Biol., 19, 2023
|
|
7OU8
 
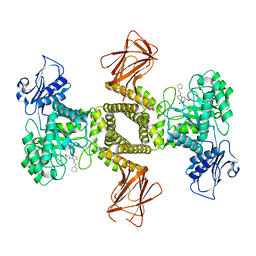 | | Human O-GlcNAc hydrolase in complex with DNJNAc-thiazolidines | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, O-GlcNAcase BT_4395, ... | | Authors: | Males, A, Davies, G.J, Gonzalez-Cuesta, M, Mellet, C.O, Fernandez, J.M.G, Sidhu, P, Ashmus, R, Busmann, J, Vocadlo, D.J, Foster, L. | | Deposit date: | 2021-06-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bicyclic Picomolar OGA Inhibitors Enable Chemoproteomic Mapping of Its Endogenous Post-translational Modifications
J.Am.Chem.Soc., 144, 2022
|
|
7EZT
 
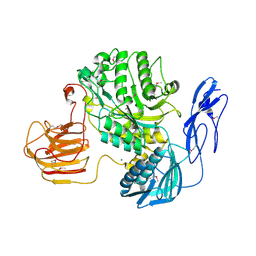 | | The structure and functional mechanism of nucleotide regulated acetylhexosaminidase Am2136 from Akkermansia muciniphila | | Descriptor: | Beta-N-acetylhexosaminidase, MAGNESIUM ION | | Authors: | Bao, R, Li, C.C, Tang, X.Y, Zhu, Y.B, Song, Y.J, Zhao, N.L, Huang, Q, Mou, X.Y, Luo, G.H, Liu, T.G. | | Deposit date: | 2021-06-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Nucleotide binding as an allosteric regulatory mechanism for Akkermansia muciniphila beta- N -acetylhexosaminidase Am2136.
Gut Microbes, 14, 2022
|
|
7DVB
 
 | | D335N variant of Bt4394 in complex with 6SO3-NAG-oxazoline intermediate | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, [(3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-2-methyl-6,7-bis(oxidanyl)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]oxazol-1-ium-5-yl]methyl sulfate | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DVA
 
 | | Structure of wild type Bt4394, a GH20 family sulfoglycosidase, in complex with 6S-GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, GLYCEROL | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DUP
 
 | | Apo structure of wild type Bt4394, a GH20 family sulfoglycosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-10 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7KHV
 
 | | CpOGA IN COMPLEX WITH LIGAND 54 | | Descriptor: | CALCIUM ION, CHLORIDE ION, N-(5-{[6-(5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)-2,6-diazaspiro[3.4]octan-2-yl]methyl}-1,3-thiazol-2-yl)acetamide, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2020-10-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Diazaspirononane Nonsaccharide Inhibitors of O-GlcNAcase (OGA) for the Treatment of Neurodegenerative Disorders.
J.Med.Chem., 63, 2020
|
|
7K41
 
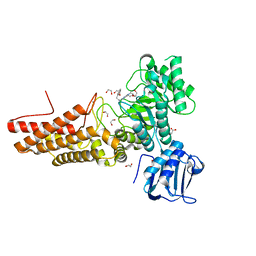 | | Bacterial O-GlcNAcase (OGA) with compound | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-methylpiperidin-1-yl)-N-(2-phenylethyl)pyrimidin-2-amine, ACETATE ION, ... | | Authors: | Lane, W, Tjhen, R, Snell, G, Sang, B. | | Deposit date: | 2020-09-14 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a Novel and Brain-Penetrant O -GlcNAcase Inhibitor via Virtual Screening, Structure-Based Analysis, and Rational Lead Optimization.
J.Med.Chem., 64, 2021
|
|
7CBO
 
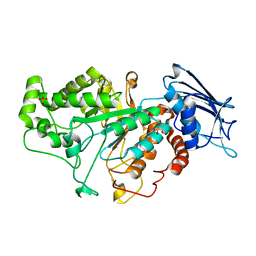 | | Crystal structure of beta-N-acetylhexosaminidase Am0868 from Akkermansia muciniphila in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, GLYCEROL, ... | | Authors: | Xu, W, Wang, M, Zhang, M. | | Deposit date: | 2020-06-13 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and biochemical analyses of beta-N-acetylhexosaminidase Am0868 from Akkermansia muciniphila involved in mucin degradation.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
7CBN
 
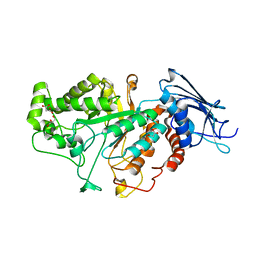 | |
6Z14
 
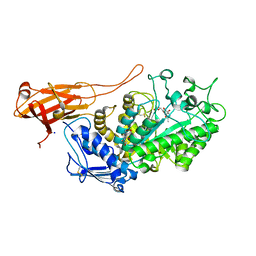 | | Structure of Bifidobacterium bifidum GH20 beta-N-beta-N-acetylhexosaminidase E553Q variant in complex with 4MU-6SGlcNAc-derived oxazoline | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-N-acetylhexosaminidase, NITRATE ION, ... | | Authors: | He, Y, Jin, Y, Rizkallah, P, Chen, P. | | Deposit date: | 2020-05-12 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure and activity of the GH20 beta-N-beta-N-acetylhexosaminidase from Bifidobacterium bifidum
To Be Published
|
|
6YXZ
 
 | |
7BWG
 
 | |
6YHH
 
 | | X-ray Structure of Flavobacterium johnsoniae chitobiase (FjGH20) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-N-acetylglucosaminidase-like protein Glycoside hydrolase family 20, GLYCEROL | | Authors: | Mazurkewich, S, Helland, R, MacKenzie, A, Eijsink, V.G.H, Pope, P.B, Branden, G, Larsbrink, J. | | Deposit date: | 2020-03-30 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights of the enzymes from the chitin utilization locus of Flavobacterium johnsoniae.
Sci Rep, 10, 2020
|
|
6PWI
 
 | | Structure of CpGH84D | | Descriptor: | 1,2-ETHANEDIOL, Putative hyaluronoglucosaminidase | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-07-23 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and functional analysis of four family 84 glycoside hydrolases from the opportunistic pathogen Clostridium perfringens.
Glycobiology, 30, 2019
|
|
6PV5
 
 | | Structure of CpGH84B | | Descriptor: | 1,2-ETHANEDIOL, Putative O-GlcNAcase nagJ | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-07-19 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural and functional analysis of four family 84 glycoside hydrolases from the opportunistic pathogen Clostridium perfringens.
Glycobiology, 30, 2019
|
|
