6PVJ
 
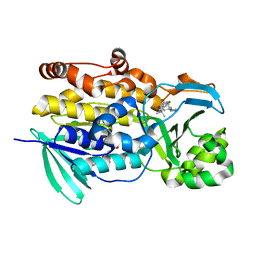 | | Crystal structure of PhqK in complex with malbrancheamide C | | Descriptor: | (5aS,12aS,13aS)-9-bromo-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
2BRY
 
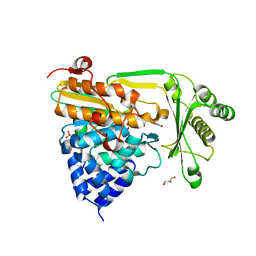 | | Crystal structure of the native monooxygenase domain of MICAL at 1.45 A resolution | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Siebold, C, Berrow, N, Walter, T.S, Harlos, K, Owens, R.J, Terman, J.R, Stuart, D.I, Kolodkin, A.L, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2005-05-13 | | Release date: | 2005-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-Resolution Structure of the Catalytic Region of Mical (Molecule Interacting with Casl), a Multidomain Flavoenzyme-Signaling Molecule.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
3ALJ
 
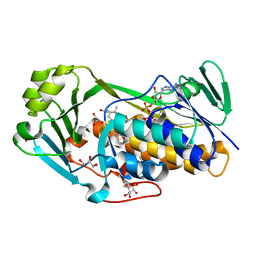 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, reduced form | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kobayashi, J, Yoshida, H, Yoshikane, Y, Kamitori, S, Yagi, T. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
5HXI
 
 | | 2-Methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5HN bound | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxypyridine-3-carboxylic acid, BETA-MERCAPTOETHANOL, ... | | Authors: | Kobayashi, J, Mikami, B. | | Deposit date: | 2016-01-30 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Role of the Tyr270 residue in 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase from Mesorhizobium loti
J. Biosci. Bioeng., 123, 2017
|
|
5NAK
 
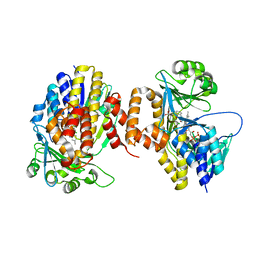 | | Pseudomonas fluorescens kynurenine 3-monooxygenase (KMO) in complex with the enzyme substrate L-kynurenine | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Taylor, M, Mowat, C.G, Rowland, P. | | Deposit date: | 2017-02-28 | | Release date: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic basis of differentiated inhibitors of the acute pancreatitis target kynurenine-3-monooxygenase.
Nat Commun, 8, 2017
|
|
4H2Q
 
 | | structure of MHPCO-5HN complex | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxypyridine-3-carboxylic acid, BETA-MERCAPTOETHANOL, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Yagi, T. | | Deposit date: | 2012-09-13 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Crystal structure of 2-Methyl-3-hydroxypyridiine-5-carboxylic acid oxygenase
To be Published
|
|
4BJZ
 
 | | Crystal structure of 3-hydroxybenzoate 6-hydroxylase uncovers lipid- assisted flavoprotein strategy for regioselective aromatic hydroxylation: Native data | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATIDYLGLYCEROL-PHOSPHOGLYCEROL, ... | | Authors: | Orru, R, Montersino, S, Barendregt, A, Westphal, A.H, van Duijn, E, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2013-04-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal Structure of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid-Assisted Flavoprotein Strategy for Regioselective Aromatic Hydroxylation
J.Biol.Chem., 288, 2013
|
|
8UIV
 
 | | H47Q NicC with bound FAD | | Descriptor: | 6-hydroxynicotinate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hicks, K.A, Perry, K. | | Deposit date: | 2023-10-10 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | Ligand bound structure of a 6-hydroxynicotinic acid 3-monooxygenase provides mechanistic insights.
Arch.Biochem.Biophys., 752, 2024
|
|
4BJY
 
 | | Crystal structure of 3-hydroxybenzoate 6-hydroxylase uncovers lipid- assisted flavoprotein strategy for regioselective aromatic hydroxylation: Platinum derivative | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATIDYLGLYCEROL-PHOSPHOGLYCEROL, ... | | Authors: | Orru, R, Montersino, S, Barendregt, A, Westphal, A.H, van Duijn, E, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2013-04-21 | | Release date: | 2013-07-24 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid-Assisted Flavoprotein Strategy for Regioselective Aromatic Hydroxylation
J.Biol.Chem., 288, 2013
|
|
7OUJ
 
 | | Crystal structure of the flavoprotein monooxygenase RubL from rubromycin biosynthesis | | Descriptor: | (2S)-hexane-1,2,6-triol, 4-HYDROXYPROLINE, CHLORIDE ION, ... | | Authors: | Saleem-Batcha, R, Toplak, M, Teufel, R. | | Deposit date: | 2021-06-11 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.573 Å) | | Cite: | Catalytic Control of Spiroketal Formation in Rubromycin Polyketide Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4GF7
 
 | | Crystal structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid oxygenase (MHPCO), unliganded form | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Sawa, Y, Yagi, T. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
5Y77
 
 | |
6JU1
 
 | | p-Hydroxybenzoate hydroxylase Y385F mutant complexed with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 4-hydroxybenzoate 3-monooxygenase, ... | | Authors: | Yato, M, Arakawa, T, Yamada, C, Fushinobu, S. | | Deposit date: | 2019-04-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Understanding the Molecular Mechanism Underlying the High Catalytic Activity ofp-Hydroxybenzoate Hydroxylase Mutants for Producing Gallic Acid.
Biochemistry, 58, 2019
|
|
3OZ2
 
 | |
7ON9
 
 | | Crystal structure of para-hydroxybenzoate-3-hydroxylase PraI | | Descriptor: | 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID | | Authors: | Zahn, M, McGeehan, J.E. | | Deposit date: | 2021-05-25 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Debottlenecking 4-hydroxybenzoate hydroxylation in Pseudomonas putida KT2440 improves muconate productivity from p-coumarate.
Metab Eng, 70, 2022
|
|
5NAB
 
 | |
6FOY
 
 | |
5NAG
 
 | |
6PVF
 
 | | Crystal structure of PhqK in complex with malbrancheamide B | | Descriptor: | (5aS,12aS,13aS)-9-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6SW2
 
 | |
1PN0
 
 | | Phenol hydroxylase from Trichosporon cutaneum | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHENOL, ... | | Authors: | Enroth, C. | | Deposit date: | 2003-06-12 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution structure of phenol hydroxylase and correction of sequence errors.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6PVG
 
 | | Crystal structure of ligand free PhqK | | Descriptor: | FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
5MZI
 
 | | Pseudomonas fluorescens kynurenine 3-monooxygenase (KMO) in complex with 3-(5-chloro-6-cyclopropoxy-2-oxo-2,3-dihydro-1,3-benzoxazol-3-yl)propanoic acid | | Descriptor: | 3-(5-chloro-6-cyclopropoxy-2-oxo-2,3-dihydro-1,3-benzoxazol-3-yl)propanoic acid, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rowland, P. | | Deposit date: | 2017-01-31 | | Release date: | 2017-04-19 | | Last modified: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Development of a Series of Kynurenine 3-Monooxygenase Inhibitors Leading to a Clinical Candidate for the Treatment of Acute Pancreatitis.
J. Med. Chem., 60, 2017
|
|
4BK1
 
 | | Crystal structure of 3-hydroxybenzoate 6-hydroxylase uncovers lipid- assisted flavoprotein strategy for regioselective aromatic hydroxylation: H213S mutant in complex with 3-hydroxybenzoate | | Descriptor: | 3-HYDROXYBENZOIC ACID, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Orru, R, Montersino, S, Barendregt, A, Westphal, A.H, van Duijn, E, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2013-04-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid-Assisted Flavoprotein Strategy for Regioselective Aromatic Hydroxylation
J.Biol.Chem., 288, 2013
|
|
6NES
 
 | | FAD-dependent monooxygenase TropB from T. stipitatus | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
