6IOZ
 
 | | Structural insights of idursulfase beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kim, H, Kim, D, Hong, J, Lee, K, Seo, J, Oh, B.H. | | Deposit date: | 2018-11-01 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights of idursulfase beta
To Be Published
|
|
5MX9
 
 | | High resolution crystal structure of the MCR-2 catalytic domain | | Descriptor: | GLYCEROL, Phosphatidylethanolamine transferase Mcr-2, ZINC ION | | Authors: | Hinchliffe, P, Coates, K, Walsh, T.R, Spencer, J. | | Deposit date: | 2017-01-22 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | 1.12 angstrom resolution crystal structure of the catalytic domain of the plasmid-mediated colistin resistance determinant MCR-2.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
3LXQ
 
 | | The Crystal Structure of a Protein in the Alkaline Phosphatase Superfamily from Vibrio parahaemolyticus to 1.95A | | Descriptor: | CHLORIDE ION, Uncharacterized protein VP1736 | | Authors: | Stein, A.J, Weger, A, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-25 | | Release date: | 2010-03-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of a Protein in the Alkaline Phosphatase Superfamily from Vibrio parahaemolyticus to 1.95A
To be Published
|
|
5AJ9
 
 | | G7 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ARYLSULFATASE, CALCIUM ION, ... | | Authors: | Miton, C.M, Fischer, G, Jonas, S, Mohammed, M.F, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2015-02-20 | | Release date: | 2016-03-16 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5WCX
 
 | |
4CXS
 
 | | G4 mutant of PAS, arylsulfatase from Pseudomonas aeruginosa, in complex with Phenylphosphonic acid | | Descriptor: | ARYLSULFATASE, CALCIUM ION, SULFATE ION, ... | | Authors: | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2014-04-08 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4KAV
 
 | |
4KAY
 
 | |
6BNC
 
 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, Thr315Ala mutant di-zinc and PEG complex | | Descriptor: | CHLORIDE ION, POLYETHYLENE GLYCOL (N=34), Phosphoethanolamine transferase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-31 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate Recognition by a Colistin Resistance Enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 13, 2018
|
|
5K4P
 
 | | Catalytic Domain of MCR-1 phosphoethanolamine transferase | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION, sorbitol | | Authors: | Stojanoski, V, Palzkill, T, Prasad, B.V.V, Sankaran, B. | | Deposit date: | 2016-05-21 | | Release date: | 2016-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.318 Å) | | Cite: | Structure of the catalytic domain of the colistin resistance enzyme MCR-1.
Bmc Biol., 14, 2016
|
|
6BNF
 
 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, mono-zinc complex | | Descriptor: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Di Leo, R, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Substrate recognition by a colistin resistance enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 2018
|
|
5LRN
 
 | | Structure of mono-zinc MCR-1 in P21 space group | | Descriptor: | GLYCEROL, Phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Hinchliffe, P, Paterson, N.G, Spencer, J. | | Deposit date: | 2016-08-19 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Insights into the Mechanistic Basis of Plasmid-Mediated Colistin Resistance from Crystal Structures of the Catalytic Domain of MCR-1.
Sci Rep, 7, 2017
|
|
6DGM
 
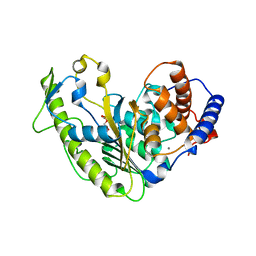 | | Streptococcus pyogenes phosphoglycerol transferase GacH in complex with sn-glycerol-1-phosphate | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Phosphoglycerol transferase GacH, ... | | Authors: | Edgar, R.J, Korotkova, N, Korotkov, K.V. | | Deposit date: | 2018-05-17 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of glycerol phosphate modification on streptococcal rhamnose polysaccharides.
Nat.Chem.Biol., 15, 2019
|
|
5LRM
 
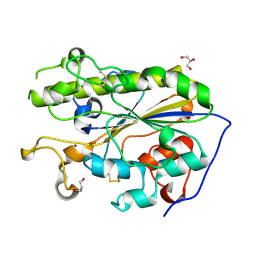 | | Structure of di-zinc MCR-1 in P41212 space group | | Descriptor: | GLYCEROL, ZINC ION, phosphatidylethanolamine transferase Mcr-1 | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2016-08-19 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into the Mechanistic Basis of Plasmid-Mediated Colistin Resistance from Crystal Structures of the Catalytic Domain of MCR-1.
Sci Rep, 7, 2017
|
|
4CYR
 
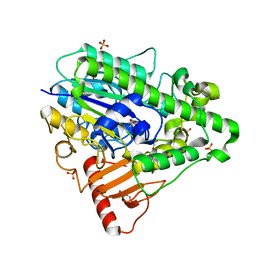 | | G4 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa | | Descriptor: | ARYLSULFATASE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2014-04-14 | | Release date: | 2015-04-29 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4CXU
 
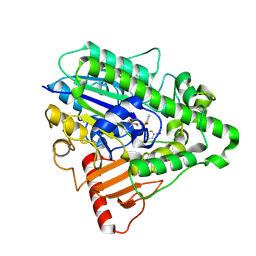 | | G4 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa, in complex with 3-Br-Phenolphenylphosphonate | | Descriptor: | 3-bromophenyl hydrogen (S)-phenylphosphonate, ARYLSULFATASE, CALCIUM ION | | Authors: | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2014-04-08 | | Release date: | 2015-04-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4CYS
 
 | | G6 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa, in complex with Phenylphosphonic acid | | Descriptor: | AMMONIUM ION, ARYLSULFATASE, CALCIUM ION, ... | | Authors: | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2014-04-14 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4CXK
 
 | | G9 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa | | Descriptor: | ARYLSULFATASE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2014-04-07 | | Release date: | 2015-04-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6LI6
 
 | | Crystal structure of MCR-1-S treated by Au(PEt3)Cl | | Descriptor: | GOLD ION, Probable phosphatidylethanolamine transferase Mcr-1, TRIETHYLPHOSPHANE | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2019-12-10 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
6LI4
 
 | | Crystal structure of MCR-1-S | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2019-12-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
6LI5
 
 | | Crystal structure of apo-MCR-1-S | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1 | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2019-12-10 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
7WAA
 
 | | Crystal structure of MCR-1-S treated by AgNO3 | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, SILVER ION | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2021-12-13 | | Release date: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Re-sensitization of mcr carrying multidrug resistant bacteria to colistin by silver.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YJP
 
 | | Crystal structure of MCR-1 treated by AuCl | | Descriptor: | GOLD ION, Probable phosphatidylethanolamine transferase Mcr-1 | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2022-07-20 | | Release date: | 2023-02-01 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Gold drugs as colistin adjuvants in the fight against MCR-1 producing bacteria.
J.Biol.Inorg.Chem., 28, 2023
|
|
7YJS
 
 | |
7YJR
 
 | |
