1WGO
 
 | | Solution structure of the PKD domain from human VPS10 domain-containing receptor SorCS2 | | Descriptor: | VPS10 domain-containing receptor SorCS2 | | Authors: | Chikayama, E, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PKD domain from human VPS10 domain-containing receptor SorCS2
To be Published
|
|
1B4R
 
 | | PKD DOMAIN 1 FROM HUMAN POLYCYSTEIN-1 | | Descriptor: | PROTEIN (PKD1_HUMAN) | | Authors: | Bycroft, M. | | Deposit date: | 1998-12-28 | | Release date: | 1999-01-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of a PKD domain from polycystin-1: implications for polycystic kidney disease.
EMBO J., 18, 1999
|
|
7KV6
 
 | |
7KV7
 
 | | Surface glycan-binding protein B from Bacteroides fluxus in complex with laminaritriose | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, LYSINE, ... | | Authors: | Tamura, K, Brumer, H, Van Petegem, F. | | Deposit date: | 2020-11-26 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Distinct protein architectures mediate species-specific beta-glucan binding and metabolism in the human gut microbiota.
J.Biol.Chem., 296, 2021
|
|
7KV5
 
 | |
5VF3
 
 | | Bacteriophage T4 isometric capsid | | Descriptor: | Capsid vertex protein gp24, Highly immunogenic outer capsid protein, Major capsid protein, ... | | Authors: | Chen, Z, Sun, L, Zhang, Z, Fokine, A, Padilla-Sanchez, V, Hanein, D, Jiang, W, Rossmann, M.G, Rao, V.B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the bacteriophage T4 isometric head at 3.3- angstrom resolution and its relevance to the assembly of icosahedral viruses.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8T9R
 
 | |
6FG9
 
 | |
8T1X
 
 | |
6FFY
 
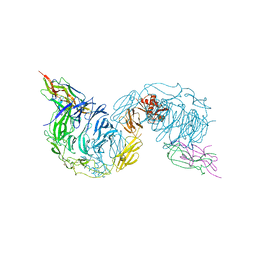 | | Structure of the mouse SorCS2-NGF complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-nerve growth factor, VPS10 domain-containing receptor SorCS2, ... | | Authors: | Leloup, N.O.L, Janssen, B.J.C. | | Deposit date: | 2018-01-09 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural insights into SorCS2-Nerve Growth Factor complex formation.
Nat Commun, 9, 2018
|
|
