2JIS
 
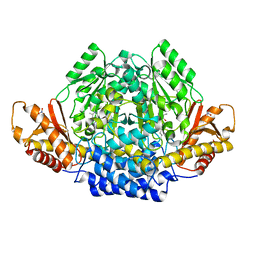 | | Human cysteine sulfinic acid decarboxylase (CSAD) in complex with PLP. | | Descriptor: | CYSTEINE SULFINIC ACID DECARBOXYLASE, NITRATE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Collins, R, Moche, M, Arrowsmith, C, Berglund, H, Busam, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Welin, M, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-30 | | Release date: | 2007-08-28 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Human Cysteine Sulfinic Acid Decarboxylase (Csad)
To be Published
|
|
6JY1
 
 | |
5HSI
 
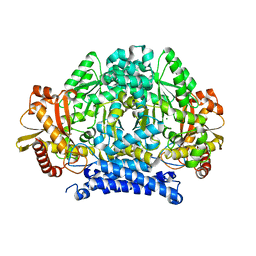 | | Crystal structure of tyrosine decarboxylase at 1.73 Angstroms resolution | | Descriptor: | MAGNESIUM ION, Putative decarboxylase | | Authors: | Ni, Y, Zhou, J, Zhu, H, Zhang, K. | | Deposit date: | 2016-01-25 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding
Sci Rep, 6, 2016
|
|
3K40
 
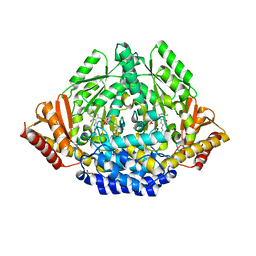 | | Crystal structure of Drosophila 3,4-dihydroxyphenylalanine decarboxylase | | Descriptor: | Aromatic-L-amino-acid decarboxylase, GLYCEROL | | Authors: | Han, Q, Ding, H, Robinson, H, Christensen, B.M, Li, J. | | Deposit date: | 2009-10-05 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and substrate specificity of Drosophila 3,4-dihydroxyphenylalanine decarboxylase
Plos One, 5, 2010
|
|
6LDR
 
 | |
4RIT
 
 | | The yellow crystal structure of pyridoxal-dependent decarboxylase from sphaerobacter thermophilus dsm 20745 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-07 | | Release date: | 2014-10-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The yellow crystal structure of pyridoxal-dependent decarboxylase from sphaerobacter thermophilus dsm 20745
To be Published
|
|
4E1O
 
 | | Human histidine decarboxylase complex with Histidine methyl ester (HME) | | Descriptor: | HISTIDINE-METHYL-ESTER, Histidine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Komori, H, Nitta, Y, Ueno, H, Higuchi, Y. | | Deposit date: | 2012-03-06 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study reveals that Ser-354 determines substrate specificity on human histidine decarboxylase
J.Biol.Chem., 287, 2012
|
|
6LDS
 
 | |
2QMA
 
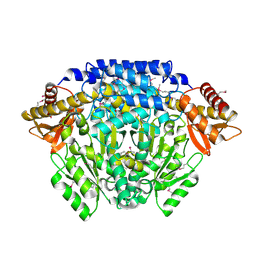 | | Crystal structure of glutamate decarboxylase domain of diaminobutyrate-pyruvate transaminase and L-2,4-diaminobutyrate decarboxylase from Vibrio parahaemolyticus | | Descriptor: | 1,2-ETHANEDIOL, Diaminobutyrate-pyruvate transaminase and L-2,4-diaminobutyrate decarboxylase | | Authors: | Osipiuk, J, Sather, A, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-14 | | Release date: | 2007-07-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray crystal structure of glutamate decarboxylase domain of diaminobutyrate-pyruvate transaminase and L-2,4-diaminobutyrate decarboxylase from Vibrio parahaemolyticus.
To be Published
|
|
8AYF
 
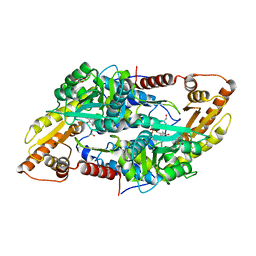 | | Crystal structure of human Sphingosine-1-phosphate lyase 1 | | Descriptor: | ACETATE ION, GLYCEROL, Sphingosine-1-phosphate lyase 1 | | Authors: | Giardina, G, Catalano, F, Pampalone, G, Cellini, B. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Dual species sphingosine-1-phosphate lyase inhibitors to combine antifungal and anti-inflammatory activities in cystic fibrosis: a feasibility study.
Sci Rep, 13, 2023
|
|
7EIX
 
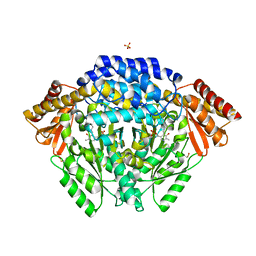 | | Human histidine decarboxylase mutant Y334F | | Descriptor: | Histidine decarboxylase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Komori, H. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of the HDC Y334F mutant
J Biol Macromol, 21, 2021
|
|
7X4Y
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase BTGAD-PLP-GABA complex | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glutamate decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Liu, S, Du, G, Wang, Y, Wen, B, Xin, F. | | Deposit date: | 2022-03-03 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
7X52
 
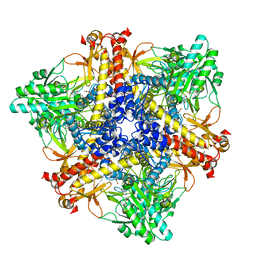 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase BTGAD-PLP complex | | Descriptor: | ACETATE ION, Glutamate decarboxylase, MALONATE ION, ... | | Authors: | Liu, S, Du, G, Wang, Y, Wen, B, Xin, F. | | Deposit date: | 2022-03-03 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
5HSJ
 
 | | Structure of tyrosine decarboxylase complex with PLP at 1.9 Angstroms resolution | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative decarboxylase | | Authors: | Ni, Y, Zhou, J, Zhu, H, Zhang, K. | | Deposit date: | 2016-01-25 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding
Sci Rep, 6, 2016
|
|
8OR9
 
 | |
2DGK
 
 | | Crystal structure of an N-terminal deletion mutant of Escherichia coli GadB in an autoinhibited state (aldamine) | | Descriptor: | 1,2-ETHANEDIOL, Glutamate decarboxylase beta, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Gruetter, M.G, Capitani, G, Gut, H. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB
Embo J., 25, 2006
|
|
4RLG
 
 | |
6LDT
 
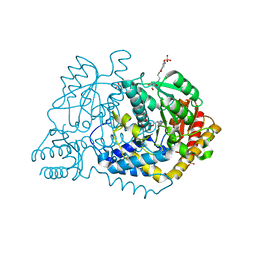 | |
2DGM
 
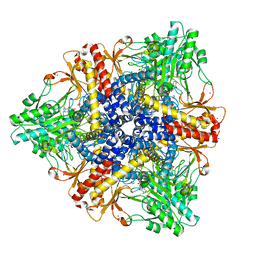 | | Crystal structure of Escherichia coli GadB in complex with iodide | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Gruetter, M.G, Capitani, G, Gut, H. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB
Embo J., 25, 2006
|
|
4RJ0
 
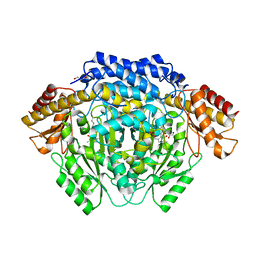 | |
6EBN
 
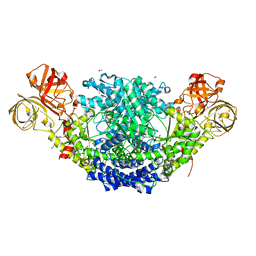 | | Crystal structure of Psilocybe cubensis noncanonical aromatic amino acid decarboxylase | | Descriptor: | FORMIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Torrens-Spence, M.P, Chun-Ting, L, Pluskal, T, Chung, Y.K, Weng, J.K. | | Deposit date: | 2018-08-06 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9663111 Å) | | Cite: | Monoamine Biosynthesis via a Noncanonical Calcium-Activatable Aromatic Amino Acid Decarboxylase in Psilocybin Mushroom.
ACS Chem. Biol., 13, 2018
|
|
6KHO
 
 | | Crystal structure of Oryza sativa TDC with PLP | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, Y.Z, Liao, L.J, Liu, X.K, Guo, Y, Zhao, Y.C, Zeng, Z.X. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-15 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Crystal structure ofOryza sativaTDC reveals the substrate specificity for TDC-mediated melatonin biosynthesis.
J Adv Res, 24, 2020
|
|
4RM1
 
 | |
6EEI
 
 | | Crystal structure of Arabidopsis thaliana phenylacetaldehyde synthase in complex with L-phenylalanine | | Descriptor: | PHENYLALANINE, SULFATE ION, Tyrosine decarboxylase 1 | | Authors: | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | Deposit date: | 2018-08-14 | | Release date: | 2018-09-19 | | Last modified: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (1.99001348 Å) | | Cite: | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7X51
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase BTGAD-PLP-GUA complex | | Descriptor: | GLUTARIC ACID, Glutamate decarboxylase, MALONATE ION, ... | | Authors: | Liu, S, Du, G, Wang, L, Wen, B, Xin, F. | | Deposit date: | 2022-03-03 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
