5M2F
 
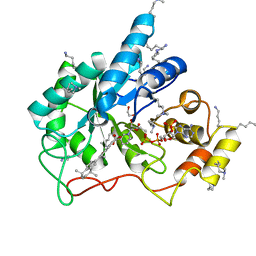 | | Crystal structure of human AKR1B10 complexed with NADP+ and the synthetic retinoid UVI2008 | | Descriptor: | 1,2-ETHANEDIOL, 3-bromo-4-[(1E)-2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)prop-1-en-1-yl]benzoic acid, Aldo-keto reductase family 1 member B10, ... | | Authors: | Ruiz, F.X, Cousido-Siah, A, Mitschler, A, Porte, S, Alvarez, S, Dominguez, M, Alvarez, R, de Lera, A.R, Pares, X, Farres, J, Podjarny, A. | | Deposit date: | 2016-10-12 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structural basis for the inhibition of AKR1B10 by the C3 brominated TTNPB derivative UVI2008.
Chem. Biol. Interact., 276, 2017
|
|
9FFD
 
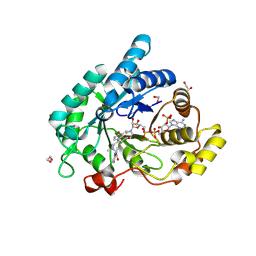 | | STRUCTURE OF ALDO-KETO REDUCTASE 1C3 (AKR1C3) IN COMPLEX WITH AN INHIBITOR MEDS765 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Aldo-keto reductase family 1 member C3, ... | | Authors: | Frydenvang, K, Hussain, S, Mirza, O.A. | | Deposit date: | 2024-05-23 | | Release date: | 2024-08-21 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | AI Based Discovery of a New AKR1C3 Inhibitor for Anticancer Applications.
Acs Med.Chem.Lett., 15, 2024
|
|
2WZT
 
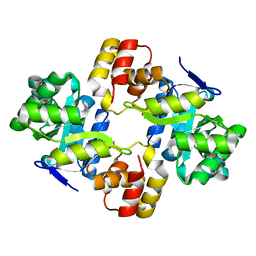 | | Crystal structure of a mycobacterium aldo-keto reductase in its apo and liganded form | | Descriptor: | ALDO-KETO REDUCTASE | | Authors: | Scoble, J, McAlister, A.D, Fulton, Z, Troy, S, Byres, E, Vivian, J.P, Brammananth, R, Wilce, M.C.J, Le Nours, J, Zaker-Tabrizi, L, Coppel, R.L, Crellin, P.K, Rossjohn, J, Beddoe, T. | | Deposit date: | 2009-12-03 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Comparative Functional Analyses of a Mycobacterium Aldo-Keto Reductase.
J.Mol.Biol., 398, 2010
|
|
8RB6
 
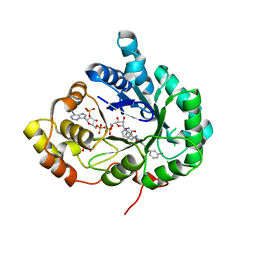 | | Structure of Aldo-Keto Reductase 1C3 (AKR1C3) in complex with an inhibitor M689, with the 3-hydroxy-benzoisoxazole moiety. Resolution 2.0A | | Descriptor: | 1,2-ETHANEDIOL, 4-[[4-(3-hydroxyphenyl)phenyl]amino]-1,2-benzoxazol-3-ol, Aldo-keto reductase family 1 member C3, ... | | Authors: | Frydenvang, K, Mirza, O.A. | | Deposit date: | 2023-12-03 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided optimization of 3-hydroxybenzoisoxazole derivatives as inhibitors of Aldo-keto reductase 1C3 (AKR1C3) to target prostate cancer.
Eur.J.Med.Chem., 268, 2024
|
|
6IJX
 
 | | Crystal Structure of AKR1C1 complexed with meclofenamic acid | | Descriptor: | 2-[(2,6-dichloro-3-methyl-phenyl)amino]benzoic acid, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X, Zhao, Y, Zhang, L, Zhang, H, Chen, Y, Hu, X. | | Deposit date: | 2018-10-12 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
3KRB
 
 | |
6XUM
 
 | | Human Aldose Reductase Mutant L300/301A in Complex with a Ligand with an IDD Structure ({5-fluoro-2-[(3-nitrobenzyl)carbamoyl]phenoxy}acetic acid) | | Descriptor: | Aldo-keto reductase family 1 member B1, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hubert, L.-S, Ley, M, Heine, A, Klebe, G. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Human Aldose Reductase Mutant L300/301A in Complex with a Ligand with an IDD Structure ({5-fluoro-2-[(3-nitrobenzyl)carbamoyl]phenoxy}acetic acid)
To Be Published
|
|
5T79
 
 | | X-Ray Crystal Structure of a Novel Aldo-keto Reductases for the Biocatalytic Conversion of 3-hydroxybutanal to 1,3-butanediol | | Descriptor: | Aldo-keto Reductase, OXIDOREDUCTASE, CHLORIDE ION, ... | | Authors: | Brunzelle, J.S, Wawrzak, Z, Evdokimova, E, Kudritska, M, Savchenko, A, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and biochemical studies of novel aldo-keto reductases for the biocatalytic conversion of 3-hydroxybutanal to 1,3-butanediol.
Appl. Environ. Microbiol., 2017
|
|
4LB4
 
 | | Crystal structure of human AR complexed with NADP+ and {2-[(4-bromo-2,3,5,6-tetrafluorobenzyl)carbamoyl]-5-chlorophenoxy}acetic acid | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {2-[(4-bromo-2,3,5,6-tetrafluorobenzyl)carbamoyl]-5-chlorophenoxy}acetic acid | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Fanfrlik, J, Kolar, M, Hobza, P, Podjarny, A. | | Deposit date: | 2013-06-20 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Modulation of aldose reductase inhibition by halogen bond tuning.
Acs Chem.Biol., 8, 2013
|
|
4LBS
 
 | | Crystal structure of human AR complexed with NADP+ and {2-[(4-bromo-2,6-difluorobenzyl)carbamoyl]-5-chlorophenoxy}acetic acid | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {2-[(4-bromo-2,6-difluorobenzyl)carbamoyl]-5-chlorophenoxy}acetic acid | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Fanfrlik, J, Kolar, M, Hobza, P, Podjarny, A. | | Deposit date: | 2013-06-21 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.76 Å) | | Cite: | Modulation of aldose reductase inhibition by halogen bond tuning.
Acs Chem.Biol., 8, 2013
|
|
1QRQ
 
 | |
1QWK
 
 | | Structural genomics of Caenorhabditis Elegans: Hypothetical 35.2 kDa protein (aldose reductase family member) | | Descriptor: | aldo-keto reductase family 1 member C1 | | Authors: | Chen, L, Zhou, X.E, Meehan, E.J, Symersky, J, Lu, S, Li, S, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-09-02 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural genomics of Caenorhabditis Elegans: Hypothetical 35.2 kDa
protein (aldose reductase family member)
To be published
|
|
8CNP
 
 | |
8CNF
 
 | |
6Y03
 
 | |
6Y1P
 
 | | Human Aldose Reductase Mutant L300/301A in Complex with a Ligand with an IDD Structure (3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid) | | Descriptor: | 3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid, Aldo-keto reductase family 1 member B1, CITRIC ACID, ... | | Authors: | Hubert, L.-S, Ley, M, Heine, A, Klebe, G. | | Deposit date: | 2020-02-13 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Human Aldose Reductase Mutant L300/301A in Complex with a Ligand with an IDD Structure (3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid)
To Be Published
|
|
7LH6
 
 | |
7MBF
 
 | | codeinone reductase isoform 1.3 Apo form | | Descriptor: | NADPH-dependent codeinone reductase 1-3 | | Authors: | Carr, S.C, Ng, K.K.S. | | Deposit date: | 2021-03-31 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of codeinone reductase reveal novel insights into aldo-keto reductase function in benzylisoquinoline alkaloid biosynthesis.
J.Biol.Chem., 297, 2021
|
|
5UXF
 
 | | Crystal Structure of mouse RECON (AKR1C13) in complex with Cyclic di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Dihydrodiol dehydrogenase | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2017-02-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Sensing of Bacterial Cyclic Dinucleotides by the Oxidoreductase RECON Promotes NF-kappa B Activation and Shapes a Proinflammatory Antibacterial State.
Immunity, 46, 2017
|
|
5DAN
 
 | |
5C7H
 
 | | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021 in complex with NADPH | | Descriptor: | Aldo-keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Seidel, R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-24 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021 in complex with NADPH
to be published
|
|
4WDT
 
 | | 17beta-HSD5 in complex with 2-nitro-5-(phenylsulfonyl)phenol | | Descriptor: | 2-nitro-5-(phenylsulfonyl)phenol, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Amano, Y, Yamaguchi, T. | | Deposit date: | 2014-09-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of complexes of type 5 17 beta-hydroxysteroid dehydrogenase with structurally diverse inhibitors: insights into the conformational changes upon inhibitor binding.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WDU
 
 | | 17beta-HSD5 in complex with 4-chloro-N-(4-chlorobenzyl)-5-nitro-1H-pyrazole-3-carboxamide | | Descriptor: | 4-chloro-N-(4-chlorobenzyl)-5-nitro-1H-pyrazole-3-carboxamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Amano, Y, Yamaguchi, T. | | Deposit date: | 2014-09-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of complexes of type 5 17 beta-hydroxysteroid dehydrogenase with structurally diverse inhibitors: insights into the conformational changes upon inhibitor binding.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WDX
 
 | | 17beta-HSD5 in complex with [4-(2-hydroxyethyl)piperidin-1-yl](5-methyl-1H-indol-2-yl)methanone | | Descriptor: | Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [4-(2-hydroxyethyl)piperidin-1-yl](5-methyl-1H-indol-2-yl)methanone | | Authors: | Amano, Y, Yamaguchi, T. | | Deposit date: | 2014-09-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structures of complexes of type 5 17 beta-hydroxysteroid dehydrogenase with structurally diverse inhibitors: insights into the conformational changes upon inhibitor binding.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6CIA
 
 | | Crystal structure of aldo-keto reductase from Klebsiella pneumoniae in complex with NADPH. | | Descriptor: | Aldo/keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lipowska, J, Leung, E.S, Shabalin, I.G, Grabowski, M, Almo, S.C, Satchell, K.J, Joachimiak, A, Lewinski, K, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of of aldo-keto reductase from Klebsiella pneumoniae in complex with NADPH.
to be published
|
|
