2HGF
 
 | | HAIRPIN LOOP CONTAINING DOMAIN OF HEPATOCYTE GROWTH FACTOR, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HEPATOCYTE GROWTH FACTOR | | Authors: | Zhou, H, Mazzulla, M.J, Kaufman, J.D, Stahl, S.J, Wingfield, P.T, Rubin, J.S, Bottaro, D.P, Byrd, R.A. | | Deposit date: | 1997-12-18 | | Release date: | 1998-06-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of hepatocyte growth factor reveals a potential heparin-binding site.
Structure, 6, 1998
|
|
1BHT
 
 | | NK1 FRAGMENT OF HUMAN HEPATOCYTE GROWTH FACTOR | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEPATOCYTE GROWTH FACTOR, SULFATE ION | | Authors: | Ultsch, M.H, Lokker, N.A, Godowski, P.J, De Vos, A.M. | | Deposit date: | 1998-06-10 | | Release date: | 1998-11-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the NK1 fragment of human hepatocyte growth factor at 2.0 A resolution.
Structure, 6, 1998
|
|
1NK1
 
 | | NK1 FRAGMENT OF HUMAN HEPATOCYTE GROWTH FACTOR/SCATTER FACTOR (HGF/SF) AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | PROTEIN (HEPATOCYTE GROWTH FACTOR PRECURSOR) | | Authors: | Chirgadze, D.Y, Hepple, J.P, Zhou, H, Byrd, R.A, Blundell, T.L, Gherardi, E. | | Deposit date: | 1998-08-20 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the NK1 fragment of HGF/SF suggests a novel mode for growth factor dimerization and receptor binding.
Nat.Struct.Biol., 6, 1999
|
|
1GMN
 
 | | CRYSTAL STRUCTURES OF NK1-HEPARIN COMPLEXES REVEAL THE BASIS FOR NK1 ACTIVITY AND ENABLE ENGINEERING OF POTENT AGONISTS OF THE MET RECEPTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEPATOCYTE GROWTH FACTOR | | Authors: | Lietha, D, Chirgadze, D.Y, Mulloy, B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2001-09-19 | | Release date: | 2001-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Nk1-Heparin Complexes Reveal the Basis for Nk1 Activity and Enable Engineering of Potent Agonists of the met Receptor
Embo J., 20, 2001
|
|
1GMO
 
 | | CRYSTAL STRUCTURES OF NK1-HEPARIN COMPLEXES REVEAL THE BASIS FOR NK1 ACTIVITY AND ENABLE ENGINEERING OF POTENT AGONISTS OF THE MET RECEPTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ... | | Authors: | Lietha, D, Chirgadze, D.Y, Mulloy, B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2001-09-20 | | Release date: | 2001-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Nk1-Heparin Complexes Reveal the Basis for Nk1 Activity and Enable Engineering of Potent Agonists of the met Receptor
Embo J., 20, 2001
|
|
1GP9
 
 | | A New Crystal Form of the Nk1 Splice Variant of Hgf/Sf Demonstrates Extensive Hinge Movement and Suggests that the Nk1 Dimer Originates by Domain Swapping | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEPATOCYTE GROWTH FACTOR | | Authors: | Watanabe, K, Chirgadze, D.Y, Lietha, D, Gherardi, E, Blundell, T.L. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A New Crystal Form of the Nk1 Splice Variant of Hgf/Sf Demonstrates Extensive Hinge Movement and Suggests that the Nk1 Dimer Originates by Domain Swapping
J.Mol.Biol., 319, 2002
|
|
1HKY
 
 | | Solution structure of a PAN module from Eimeria tenella | | Descriptor: | MICRONEME PROTEIN 5 PRECURSOR | | Authors: | Brown, P.J, Mulvey, D, Potts, J.R, Tomley, F.M, Campbell, I.D. | | Deposit date: | 2002-10-03 | | Release date: | 2002-10-17 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Pan Module from the Apicomplexan Parasite Eimeria Tenella
J.Struct.Funct.Genom., 4, 2003
|
|
2J8J
 
 | | Solution Structure of the A4 Domain of Blood Coagulation Factor XI | | Descriptor: | COAGULATION FACTOR XI | | Authors: | Samuel, D, Cheng, H, Riley, P.W, Canutescu, A.A, Bu, Z, Walsh, P.N, Roder, H. | | Deposit date: | 2006-10-25 | | Release date: | 2007-10-02 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the A4 Domain of Factor Xi Sheds Light on the Mechanism of Zymogen Activation.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2J8L
 
 | | FXI Apple 4 domain loop-out conformation | | Descriptor: | COAGULATION FACTOR XI | | Authors: | Samuel, D, Cheng, H, Riley, P.W, Canutescu, A.A, Bu, Z, Walsh, P.N, Roder, H. | | Deposit date: | 2006-10-25 | | Release date: | 2007-10-02 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the A4 Domain of Factor Xi Sheds Light on the Mechanism of Zymogen Activation.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2QJ2
 
 | | A Mechanistic Basis for Converting a Receptor Tyrosine Kinase Agonist to an Antagonist | | Descriptor: | Hepatocyte growth factor, SULFATE ION | | Authors: | Tolbert, W.D, Daugherty, J, Gao, C.-F, Xe, Q, Miranti, C, Gherardi, E, Vande Woude, G, Xu, H.E. | | Deposit date: | 2007-07-06 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A mechanistic basis for converting a receptor tyrosine kinase agonist to an antagonist
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2QJ4
 
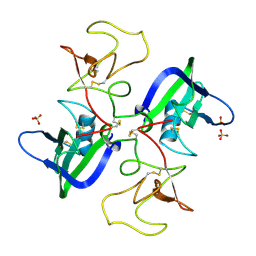 | | A Mechanistic Basis for Converting a Receptor Tyrosine Kinase Agonist to an Antagonist | | Descriptor: | Hepatocyte growth factor, SULFATE ION | | Authors: | Tolbert, W.D, Daugherty, J, Gao, C.-F, Xe, Q, Miranti, C, Gherardi, E, Vande Woude, G, Xu, H.E. | | Deposit date: | 2007-07-06 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A mechanistic basis for converting a receptor tyrosine kinase agonist to an antagonist
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3HMS
 
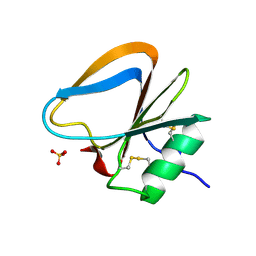 | |
3HMT
 
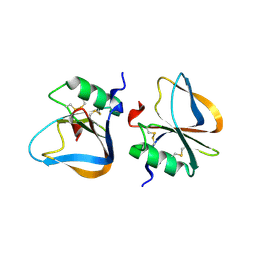 | |
3HMR
 
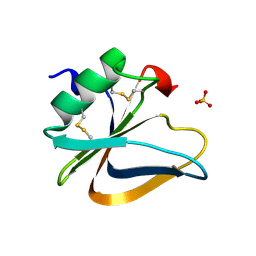 | |
3HN4
 
 | |
3MKP
 
 | | Crystal structure of 1K1 mutant of Hepatocyte Growth Factor/Scatter Factor fragment NK1 in complex with heparin | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Gherardi, E, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Engineering a fragment of Hepatocyte Growth Factor/Scatter Factor for tissue and organ regeneration
To be Published
|
|
2YIL
 
 | | Crystal Structure of Parasite Sarcocystis muris Lectin SML-2 | | Descriptor: | CHLORIDE ION, GLYCEROL, MICRONEME ANTIGEN L2, ... | | Authors: | Mueller, J.J, Weiss, M.S, Heinemann, U. | | Deposit date: | 2011-05-16 | | Release date: | 2011-11-02 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pan-Modular Structure of Microneme Protein Sml-2 from Parasite Sarcocystis Muris at 1.95 A Resolution and its Complex with 1-Thio-Beta-D-Galactose.
Acta Crystallogr.,Sect.D, D67, 2011
|
|
2YIP
 
 | |
2YIO
 
 | |
3SP8
 
 | | Crystal structure of NK2 in complex with fractionated Heparin DP10 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Recacha, R, Mulloy, B, Gherardi, E. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of NK2 in complex with fractionated Heparin DP10
TO BE PUBLISHED
|
|
2LL4
 
 | |
2LL3
 
 | |
4A5T
 
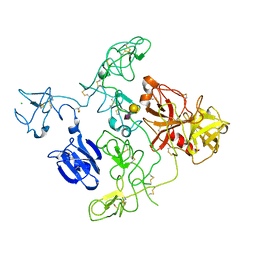 | | STRUCTURAL BASIS FOR THE CONFORMATIONAL MODULATION | | Descriptor: | CHLORIDE ION, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, PLASMINOGEN | | Authors: | Xue, Y, Bodin, C, Olsson, K. | | Deposit date: | 2011-10-28 | | Release date: | 2012-05-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Crystal Structure of the Native Plasminogen Reveals an Activation-Resistant Compact Conformation.
J. Thromb. Haemost., 10, 2012
|
|
4A5V
 
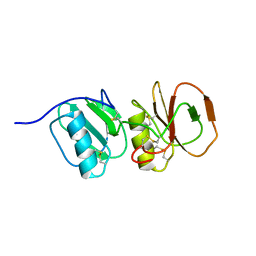 | | Solution structure ensemble of the two N-terminal apple domains (residues 58-231) of Toxoplasma gondii microneme protein 4 | | Descriptor: | MICRONEMAL PROTEIN 4 | | Authors: | Marchant, J, Cowper, B, Liu, Y, Lai, L, Pinzan, C, Marq, J.B, Friedrich, N, Sawmynaden, K, Chai, W, Childs, R.A, Saouros, S, Simpson, P, Barreira, M.C.R, Feizi, T, Soldati-Favre, D, Matthews, S. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Galactose Recognition by the Apicomplexan Parasite Toxoplasma Gondii.
J.Biol.Chem., 287, 2012
|
|
4DUR
 
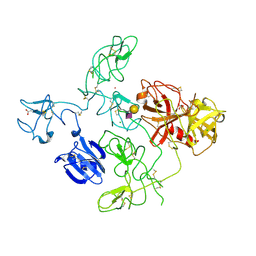 | | The X-ray Crystal Structure of Full-Length type II Human Plasminogen | | Descriptor: | ACETATE ION, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Law, R.H.P, Caradoc-Davies, T, Whisstock, J.C. | | Deposit date: | 2012-02-22 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The X-ray crystal structure of full-length human plasminogen
Cell Rep, 1, 2012
|
|
