2A65
 
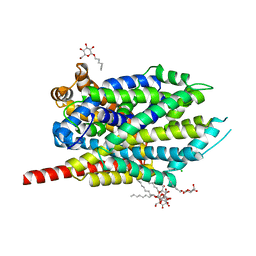 | | Crystal structure of LEUTAA, a bacterial homolog of Na+/Cl--dependent neurotransmitter transporters | | Descriptor: | CHLORIDE ION, LEUCINE, Na(+):neurotransmitter symporter (Snf family), ... | | Authors: | Yamashita, A, Singh, S.K, Kawate, T, Jin, Y, Gouaux, E. | | Deposit date: | 2005-07-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a bacterial homologue of Na(+)/Cl(-)-dependent neurotransmitter transporters.
Nature, 437, 2005
|
|
7AHK
 
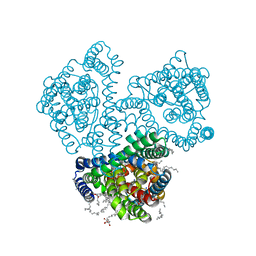 | | Crystal structure of the outward-facing state of the substrate-free Na+-only bound glutamate transporter homolog GltPh | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, GLYCEROL, ... | | Authors: | Kovalev, K, Alleva, C, Astashkin, A, Machtens, J.-P, Fahlke, C, Gordeliy, V. | | Deposit date: | 2020-09-24 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Na + -dependent gate dynamics and electrostatic attraction ensure substrate coupling in glutamate transporters.
Sci Adv, 6, 2020
|
|
8ACY
 
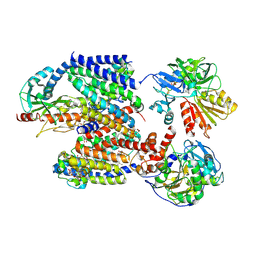 | | X-ray structure of Na+-NQR from Vibrio cholerae at 3.5 A resolution | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Fritz, G. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4HQJ
 
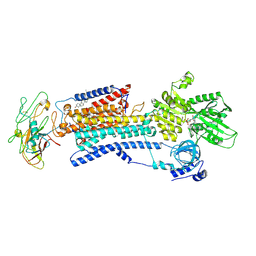 | | Crystal structure of Na+,K+-ATPase in the Na+-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Nyblom, M, Reinhard, L, Gourdon, P, Nissen, P. | | Deposit date: | 2012-10-25 | | Release date: | 2013-10-02 | | Last modified: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Crystal structure of Na+, K(+)-ATPase in the Na(+)-bound state.
Science, 342, 2013
|
|
7WYU
 
 | | Cryo-EM structure of Na+,K+-ATPase in the E2P state formed by ATP | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-02-16 | | Release date: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryoelectron microscopy of Na + ,K + -ATPase in the two E2P states with and without cardiotonic steroids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WYX
 
 | | Cryo-EM structure of Na+,K+-ATPase in the E2P state formed by ATP with istaroxime | | Descriptor: | (3E,5S,8R,9S,10R,13S,14S)-3-(2-azanylethoxyimino)-10,13-dimethyl-1,2,4,5,7,8,9,11,12,14,15,16-dodecahydrocyclopenta[a]phenanthrene-6,17-dione, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-02-16 | | Release date: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryoelectron microscopy of Na + ,K + -ATPase in the two E2P states with and without cardiotonic steroids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WYV
 
 | | Cryo-EM structure of Na+,K+-ATPase in the E2P state formed by ATP in the presence of 40 mM Mg2+ | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-02-16 | | Release date: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryoelectron microscopy of Na + ,K + -ATPase in the two E2P states with and without cardiotonic steroids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WYZ
 
 | | Cryo-EM structure of Na+,K+-ATPase in the E2P state formed by ATP with ouabain | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-02-16 | | Release date: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryoelectron microscopy of Na + ,K + -ATPase in the two E2P states with and without cardiotonic steroids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6QK8
 
 | |
4PV3
 
 | | Crystal structure of potassium-dependent plant-type L-asparaginase from Phaseolus vulgaris in complex with Na+ cations | | Descriptor: | L-ASPARAGINASE ALPHA SUBUNIT, L-ASPARAGINASE BETA SUBUNIT, SODIUM ION | | Authors: | Bejger, M, Gilski, M, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2014-03-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Na+/K+ exchange switches the catalytic apparatus of potassium-dependent plant L-asparaginase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PV2
 
 | | Crystal structure of potassium-dependent plant-type L-asparaginase from Phaseolus vulgaris in complex with K+ and Na+ cations | | Descriptor: | L-ASPARAGINASE ALPHA SUBUNIT, L-ASPARAGINASE BETA SUBUNIT, NITRATE ION, ... | | Authors: | Bejger, M, Gilski, M, Imiolczyk, B, Clavel, D, Jaskolski, M. | | Deposit date: | 2014-03-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Na+/K+ exchange switches the catalytic apparatus of potassium-dependent plant L-asparaginase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2AHY
 
 | | Na+ complex of the NaK Channel | | Descriptor: | CALCIUM ION, Potassium channel protein, SODIUM ION | | Authors: | Shi, N, Ye, S, Alam, A, Chen, L, Jiang, Y. | | Deposit date: | 2005-07-28 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic structure of a Na+- and K+-conducting channel.
Nature, 440, 2006
|
|
8JFZ
 
 | | Cryo-EM structure of Na+,K+-ATPase in the E1.Mg2+ state. | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Vilsen, B, Cornelius, F, Toyoshima, C. | | Deposit date: | 2023-05-19 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Crystal structures of Na + ,K + -ATPase reveal the mechanism that converts the K + -bound form to Na + -bound form and opens and closes the cytoplasmic gate.
Febs Lett., 597, 2023
|
|
8JBK
 
 | | Crystal structure of Na+,K+-ATPase in the E1.3Na+ state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Vilsen, B, Cornelius, F, Toyoshima, C. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Na + ,K + -ATPase reveal the mechanism that converts the K + -bound form to Na + -bound form and opens and closes the cytoplasmic gate.
Febs Lett., 597, 2023
|
|
8K1L
 
 | | Cryo-EM structure of Na+,K+-ATPase alpha2 from Artemia salina in cation-free E2P form | | Descriptor: | Na+,K+-ATPase alpha2KK, Na+,K+-ATPase beta2, TETRAFLUOROALUMINATE ION | | Authors: | Abe, K, Artigas, P. | | Deposit date: | 2023-07-11 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | A Na pump with reduced stoichiometry is up-regulated by brine shrimp in extreme salinities.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5UL7
 
 | |
5ULE
 
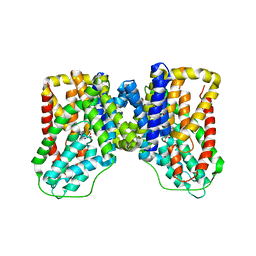 | |
8T60
 
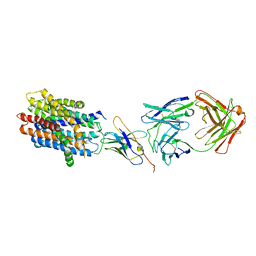 | |
7Y45
 
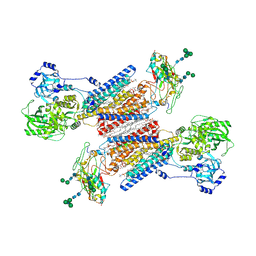 | | Cryo-EM structure of the Na+,K+-ATPase in the E2.2K+ state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-06-14 | | Release date: | 2022-07-13 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-electron microscopy of Na + ,K + -ATPase reveals how the extracellular gate locks in the E2·2K + state.
Febs Lett., 596, 2022
|
|
7Y46
 
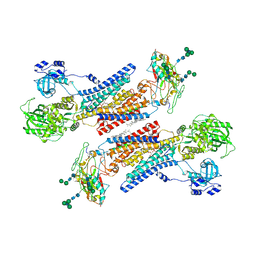 | | Cryo-EM structure of the Na+,K+-ATPase in the E2.2K+ state after addition of ATP | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-06-14 | | Release date: | 2022-07-13 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Cryo-electron microscopy of Na + ,K + -ATPase reveals how the extracellular gate locks in the E2·2K + state.
Febs Lett., 596, 2022
|
|
4HYT
 
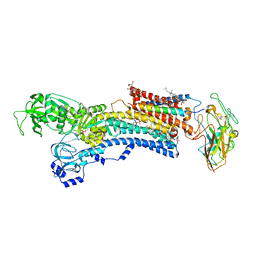 | | Na,K-ATPase in the E2P state with bound ouabain and Mg2+ in the cation-binding site | | Descriptor: | 1-O-decanoyl-beta-D-tagatofuranosyl beta-D-allopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Laursen, M, Yatime, L, Nissen, P, Fedosova, N.U. | | Deposit date: | 2012-11-14 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Crystal structure of the high-affinity Na+,K+-ATPase-ouabain complex with Mg2+ bound in the cation binding site.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3BEU
 
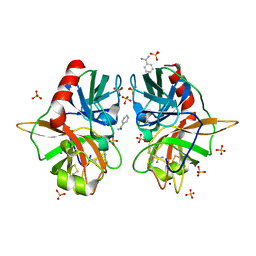 | | Na+-Dependent Allostery Mediates Coagulation Factor Protease Active Site Selectivity | | Descriptor: | BENZAMIDINE, CALCIUM ION, SODIUM ION, ... | | Authors: | Page, M.J, Carrell, C.J, Di Cera, E. | | Deposit date: | 2007-11-20 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Engineering protein allostery: 1.05 A resolution structure and enzymatic properties of a Na+-activated trypsin.
J.Mol.Biol., 378, 2008
|
|
1ZCD
 
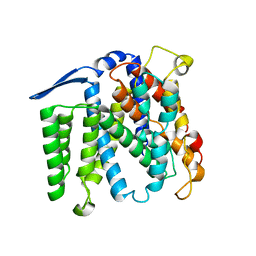 | | Crystal structure of the Na+/H+ antiporter NhaA | | Descriptor: | Na(+)/H(+) antiporter 1 | | Authors: | Hunte, C, Screpanti, E, Venturi, M, Rimon, A, Padan, E, Michel, H. | | Deposit date: | 2005-04-11 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH.
Nature, 435, 2005
|
|
2A2Q
 
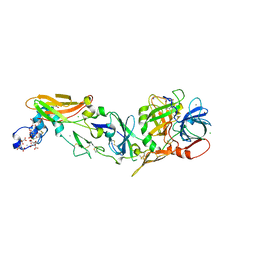 | | Complex of Active-site Inhibited Human Coagulation Factor VIIa with Human Soluble Tissue Factor in the Presence of Ca2+, Mg2+, Na+, and Zn2+ | | Descriptor: | CALCIUM ION, CHLORIDE ION, Coagulation factor VII, ... | | Authors: | Bajaj, S.P, Bajaj, M, Schmidt, A.E, Padmanabhan, K. | | Deposit date: | 2005-06-22 | | Release date: | 2006-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structures of p-aminobenzamidine- and benzamidine-VIIa/soluble tissue factor: unpredicted conformation of the 192-193 peptide bond and mapping of Ca2+, Mg2+, Na+, and Zn2+ sites in factor VIIa.
J.Biol.Chem., 281, 2006
|
|
8IJM
 
 | | Cyo-EM structure of K794A non-gastric proton pump in Na+ bound E1AMPPCP state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Abe, K. | | Deposit date: | 2023-02-27 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | An unusual conformation from Na + -sensitive non-gastric proton pump mutants reveals molecular mechanisms of cooperative Na + -binding.
Biochim Biophys Acta Mol Cell Res, 1870, 2023
|
|
