1F8A
 
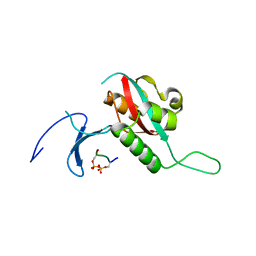 | | STRUCTURAL BASIS FOR THE PHOSPHOSERINE-PROLINE RECOGNITION BY GROUP IV WW DOMAINS | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1, Y(SEP)PT(SEP)S PEPTIDE | | Authors: | Verdecia, M.A, Bowman, M.E, Lu, K.P, Hunter, T, Noel, J.P. | | Deposit date: | 2000-06-29 | | Release date: | 2000-08-23 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for phosphoserine-proline recognition by group IV WW domains.
Nat.Struct.Biol., 7, 2000
|
|
3OOB
 
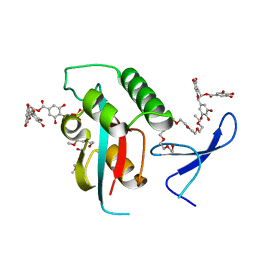 | | Structural and functional insights of directly targeting Pin1 by Epigallocatechin-3-gallate | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Urusova, D.V, Shim, J.-H, Kim, D.-J, Jung, S.K, Zykova, T.A, Bode, A.M, Dong, Z. | | Deposit date: | 2010-08-30 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Epigallocatechin-gallate suppresses tumorigenesis by directly targeting Pin1.
Cancer Prev Res (Phila), 4, 2011
|
|
4QIB
 
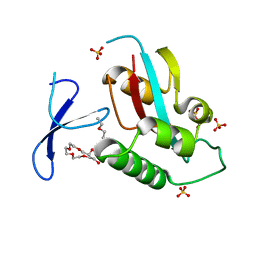 | | Oxidation-Mediated Inhibition of the Peptidyl-Prolyl Isomerase Pin1 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION | | Authors: | Innes, B.T, Sowole, M.A, Konermann, L, Litchfield, D.W, Brandl, C.J, Shilton, B.H. | | Deposit date: | 2014-05-30 | | Release date: | 2015-02-04 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (1.865 Å) | | Cite: | Peroxide-mediated oxidation and inhibition of the peptidyl-prolyl isomerase Pin1.
Biochim.Biophys.Acta, 1852, 2015
|
|
3WH0
 
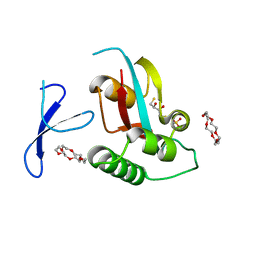 | | Structure of Pin1 Complex with 18-crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Lee, C.C, Liu, C.I, Jeng, W.Y, Wang, A.H.J. | | Deposit date: | 2013-08-20 | | Release date: | 2014-10-15 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crowning proteins: modulating the protein surface properties using crown ethers.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4U85
 
 | | Human Pin1 with cysteine sulfinic acid 113 | | Descriptor: | POLYETHYLENE GLYCOL (N=34), Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Li, W, Zhang, Y. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer's disease.
Neurobiol.Dis., 76, 2015
|
|
4U86
 
 | | Human Pin1 with cysteine sulfonic acid 113 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Li, W, Zhang, Y. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer's disease.
Neurobiol.Dis., 76, 2015
|
|
4U84
 
 | | Human Pin1 with S-hydroxyl-cysteine 113 | | Descriptor: | POLYETHYLENE GLYCOL (N=34), Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Li, W, Zhang, Y. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer's disease.
Neurobiol.Dis., 76, 2015
|
|
7EKV
 
 | | Crystal Structure of human Pin1 complexed with a covalent inhibitor | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-((5-phenylfuran-2-yl)methyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7EFJ
 
 | | Crystal Structure Analysis of human PIN1 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-(furan-2-ylmethyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-03-21 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7EFX
 
 | | Crystal Structure of human PIN1 complexed with covalent inhibitor | | Descriptor: | 4-((5-bromofuran-2-yl)methyl)-8-(2-chloroacetyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J, Zhu, R, Pei, Y. | | Deposit date: | 2021-03-23 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
5B3Y
 
 | |
7F0M
 
 | | Crystal Structure of human Pin1 complexed with a potent covalent inhibitor | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 8-(2-chloranylethanoyl)-4-[(5-naphthalen-1-ylfuran-2-yl)methyl]-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-06-05 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
5B3Z
 
 | |
5BMY
 
 | |
7KKF
 
 | |
5TJ8
 
 | |
5TJ7
 
 | |
5TJQ
 
 | | Structure of WWP2 2,3-linker-HECT | | Descriptor: | NEDD4-like E3 ubiquitin-protein ligase WWP2,NEDD4-like E3 ubiquitin-protein ligase WWP2 | | Authors: | Chen, Z, Gabelli, S.B. | | Deposit date: | 2016-10-04 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A Tunable Brake for HECT Ubiquitin Ligases.
Mol. Cell, 66, 2017
|
|
5UY9
 
 | | Prolyl isomerase Pin1 R14A mutant bound with Brd4 peptide | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Brd4 peptide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Dong, S.-H, Nair, S. | | Deposit date: | 2017-02-23 | | Release date: | 2017-04-26 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Prolyl isomerase PIN1 regulates the stability, transcriptional activity and oncogenic potential of BRD4.
Oncogene, 36, 2017
|
|
3KAF
 
 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 3-(1H-benzimidazol-2-yl)-N-(1-benzothiophen-2-ylcarbonyl)-D-alanine, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KAH
 
 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 3-(1H-benzimidazol-2-yl)-N-[(1-methyl-3-phenyl-1H-pyrazol-5-yl)carbonyl]-D-alanine, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KAI
 
 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | (2R)-2-[(2-methyl-5-phenyl-pyrazol-3-yl)carbonylamino]-3-naphthalen-2-yl-propanoic acid, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KAD
 
 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 3-(1H-benzimidazol-2-yl)-N-(3-phenylpropanoyl)-D-alanine, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KAB
 
 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 6-methyl-1H-indole-2-carboxylic acid, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KAG
 
 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 3-(1H-benzimidazol-2-yl)-N-[(2-methylfuran-3-yl)carbonyl]-D-alanine, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
