6ACQ
 
 | |
1F12
 
 | | L-3-HYDROXYACYL-COA DEHYDROGENASE COMPLEXED WITH 3-HYDROXYBUTYRYL-COA | | Descriptor: | 3-HYDROXYBUTANOYL-COENZYME A, L-3-HYDROXYACYL-COA DEHYDROGENASE | | Authors: | Barycki, J.J, O'Brien, L.K, Strauss, A.W, Banaszak, L.J. | | Deposit date: | 2000-05-18 | | Release date: | 2000-09-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sequestration of the active site by interdomain shifting. Crystallographic and spectroscopic evidence for distinct conformations of L-3-hydroxyacyl-CoA dehydrogenase.
J.Biol.Chem., 275, 2000
|
|
1F14
 
 | |
1F0Y
 
 | | L-3-HYDROXYACYL-COA DEHYDROGENASE COMPLEXED WITH ACETOACETYL-COA AND NAD+ | | Descriptor: | ACETOACETYL-COENZYME A, L-3-HYDROXYACYL-COA DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Barycki, J.J, O'Brien, L.K, Strauss, A.W, Banaszak, L.J. | | Deposit date: | 2000-05-17 | | Release date: | 2000-09-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sequestration of the active site by interdomain shifting. Crystallographic and spectroscopic evidence for distinct conformations of L-3-hydroxyacyl-CoA dehydrogenase.
J.Biol.Chem., 275, 2000
|
|
1F17
 
 | | L-3-HYDROXYACYL-COA DEHYDROGENASE COMPLEXED WITH NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-3-HYDROXYACYL-COA DEHYDROGENASE | | Authors: | Barycki, J.J, O'Brien, L.K, Strauss, A.W, Banaszak, L.J. | | Deposit date: | 2000-05-18 | | Release date: | 2000-09-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sequestration of the active site by interdomain shifting. Crystallographic and spectroscopic evidence for distinct conformations of L-3-hydroxyacyl-CoA dehydrogenase.
J.Biol.Chem., 275, 2000
|
|
4J0E
 
 | |
4J0F
 
 | |
1ZCJ
 
 | |
1ZEJ
 
 | | Crystal structure of the 3-hydroxyacyl-coa dehydrogenase (hbd-9, af2017) from archaeoglobus fulgidus dsm 4304 at 2.00 A resolution | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 3-hydroxyacyl-CoA dehydrogenase, CHLORIDE ION | | Authors: | Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2005-04-18 | | Release date: | 2005-05-03 | | Last modified: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 3-hydroxyacyl-CoA dehydrogenase (HBD-9) (np_070841.1) from Archaeoglobus fulgidus at 2.00 A resolution
To be published
|
|
4KUH
 
 | |
4KUE
 
 | |
4KUG
 
 | |
5MGB
 
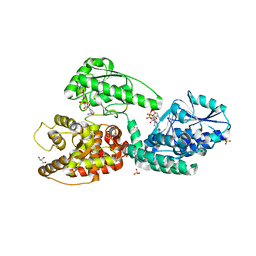 | | Crystal Structure of Rat Peroxisomal Multifunctional enzyme Type-1 (RPMFE1) Complexed with Acetoacetyl-CoA and NAD | | Descriptor: | ACETOACETYL-COENZYME A, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kasaragod, P, Kiema, T.-R, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2016-11-21 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural enzymology comparisons of multifunctional enzyme, type-1 (MFE1): the flexibility of its dehydrogenase part.
FEBS Open Bio, 7, 2017
|
|
6TNM
 
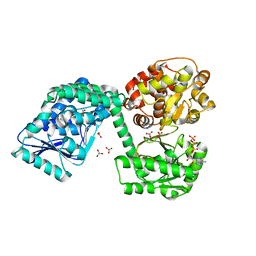 | | E. coli aerobic trifunctional enzyme subunit-alpha | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Fatty acid oxidation complex subunit alpha, GLYCEROL, ... | | Authors: | Sah-Teli, S.K, Hynonen, M.J, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2019-12-09 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Insights into the stability and substrate specificity of the E. coli aerobic beta-oxidation trifunctional enzyme complex.
J.Struct.Biol., 210, 2020
|
|
5OMO
 
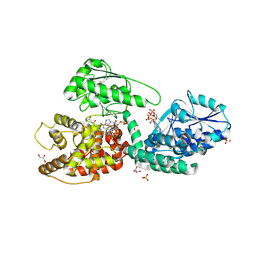 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE-1 (RPMFE1) COMPLEXED WITH WITH 3S-HYDROXY-DECANOYL-COA AND 3-KETO-DECANOYL-COA | | Descriptor: | (S)-3-HYDROXYDECANOYL-COA, 3-KETO-DECANOYL-COA, GLYCEROL, ... | | Authors: | Kasaragod, P, Kiema, T.-R, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME
TYPE 1 (RPMFE1) COMPLEXED WITH 3S-HYDROXY-DECANOYL-COA AND 3-
KETO-DECANOYL-COA
Not Published
|
|
6DV2
 
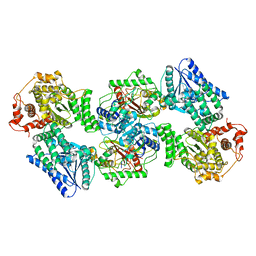 | | Crystal Structure of Human Mitochondrial Trifunctional Protein | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Fu, Z, Xia, C, Battaile, K.P, Kim, J.P. | | Deposit date: | 2018-06-22 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of human mitochondrial trifunctional protein, a fatty acid beta-oxidation metabolon.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
2D3T
 
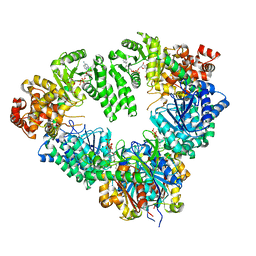 | | Fatty Acid beta-oxidation multienzyme complex from Pseudomonas Fragi, Form V | | Descriptor: | 3-ketoacyl-CoA thiolase, ACETYL COENZYME *A, Fatty oxidation complex alpha subunit, ... | | Authors: | Tsuchiya, D, Shimizu, N, Ishikawa, M, Suzuki, Y, Morikawa, K. | | Deposit date: | 2005-10-01 | | Release date: | 2006-02-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ligand-Induced Domain Rearrangement of Fatty Acid beta-Oxidation Multienzyme Complex
Structure, 14, 2006
|
|
6IUN
 
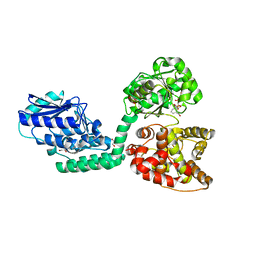 | |
6IUM
 
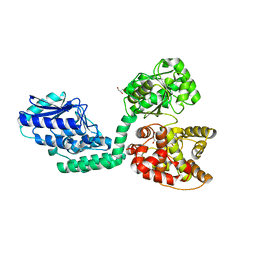 | |
2X58
 
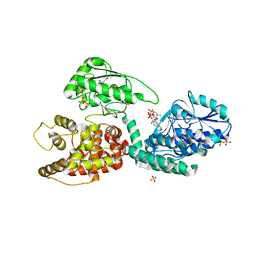 | | The crystal structure of MFE1 liganded with CoA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COENZYME A, GLYCEROL, ... | | Authors: | Kasaragod, P, Venkatesan, R, Kiema, T.R, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2010-02-05 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of Liganded Rat Peroxisomal Multifunctional Enzyme Type 1: A Flexible Molecule with Two Interconnected Active Sites
J.Biol.Chem., 285, 2010
|
|
2WTB
 
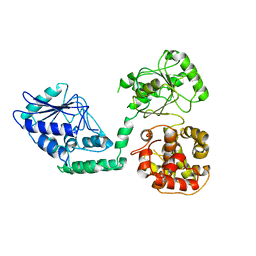 | | Arabidopsis thaliana multifuctional protein, MFP2 | | Descriptor: | FATTY ACID MULTIFUNCTIONAL PROTEIN (ATMFP2) | | Authors: | Arent, S, Pye, V.E, Christensen, C.E, Norgaard, A, Henriksen, A. | | Deposit date: | 2009-09-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Multi-Functional Protein in Peroxisomal Beta-Oxidation. Structure and Substrate Specificity of the Arabidopsis Thaliana Protein, Mfp2
J.Biol.Chem., 285, 2010
|
|
8BRJ
 
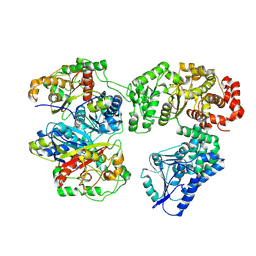 | | Escherichia coli anaerobic fatty acid beta oxidation trifunctional enzyme (anEcTFE) trimeric complex | | Descriptor: | 3-ketoacyl-CoA thiolase FadI, Fatty acid oxidation complex subunit alpha | | Authors: | Sah-Teli, S.K, Pinkas, M, Novacek, J, Venkatesan, R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes.
Structure, 31, 2023
|
|
8BNU
 
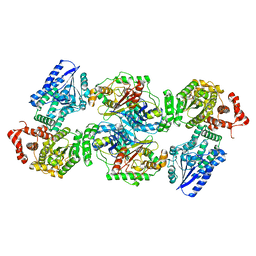 | | Escherichia coli anaerobic fatty acid beta oxidation trifunctional enzyme (anEcTFE) tetrameric complex | | Descriptor: | 3-ketoacyl-CoA thiolase FadI, Fatty acid oxidation complex subunit alpha | | Authors: | Sah-Teli, S.K, Pinkas, M, Novacek, J, Venkatesan, R. | | Deposit date: | 2022-11-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes.
Structure, 31, 2023
|
|
8BNR
 
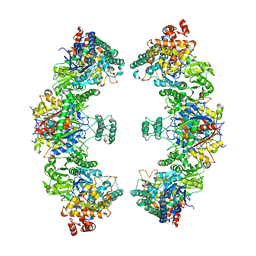 | | Escherichia coli anaerobic fatty acid beta oxidation trifunctional enzyme (anEcTFE) octameric complex | | Descriptor: | 3-ketoacyl-CoA thiolase FadI, Fatty acid oxidation complex subunit alpha | | Authors: | Sah-Teli, S.K, Pinkas, M, Novacek, J, Venkatesan, R. | | Deposit date: | 2022-11-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (10.3 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes.
Structure, 31, 2023
|
|
5ZQZ
 
 | | Structure of human mitochondrial trifunctional protein, tetramer | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Liang, K, Li, N, Dai, J, Wang, X, Liu, P, Chen, X, Wang, C, Gao, N, Xiao, J. | | Deposit date: | 2018-04-20 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of human mitochondrial trifunctional protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
