1AI3
 
 | | ORBITAL STEERING IN THE CATALYTIC POWER OF ENZYMES: SMALL STRUCTURAL CHANGES WITH LARGE CATALYTIC CONSEQUENCES | | Descriptor: | ISOCITRATE DEHYDROGENASE, ISOCITRIC ACID, MAGNESIUM ION, ... | | Authors: | Stoddard, B.L, Mesecar, A, Koshland Junior, D.E. | | Deposit date: | 1997-04-30 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Orbital steering in the catalytic power of enzymes: small structural changes with large catalytic consequences.
Science, 277, 1997
|
|
1SJS
 
 | |
2AYQ
 
 | |
1A05
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 3-ISOPROPYLMALATE DEHYDROGENASE FROM THIOBACILLUS FERROOXIDANS WITH 3-ISOPROPYLMALATE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, 3-ISOPROPYLMALIC ACID, MAGNESIUM ION | | Authors: | Imada, K, Inagaki, K, Matsunami, H, Kawaguchi, H, Tanaka, H, Tanaka, N, Namba, K. | | Deposit date: | 1997-12-09 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of 3-isopropylmalate dehydrogenase in complex with 3-isopropylmalate at 2.0 A resolution: the role of Glu88 in the unique substrate-recognition mechanism.
Structure, 6, 1998
|
|
1BL5
 
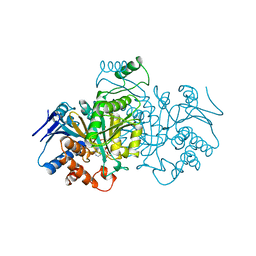 | | ISOCITRATE DEHYDROGENASE FROM E. COLI SINGLE TURNOVER LAUE STRUCTURE OF RATE-LIMITED PRODUCT COMPLEX, 10 MSEC TIME RESOLUTION | | Descriptor: | 2-OXOGLUTARIC ACID, ISOCITRATE DEHYDROGENASE, MAGNESIUM ION, ... | | Authors: | Stoddard, B.L, Cohen, B, Brubaker, M, Mesecar, A, Koshland Junior, D.E. | | Deposit date: | 1998-07-23 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Millisecond Laue structures of an enzyme-product complex using photocaged substrate analogs.
Nat.Struct.Biol., 5, 1998
|
|
1WAL
 
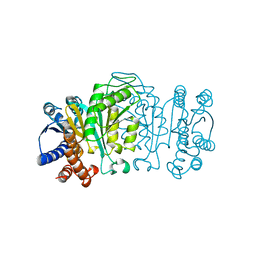 | | 3-ISOPROPYLMALATE DEHYDROGENASE (IPMDH) MUTANT (M219A)FROM THERMUS THERMOPHILUS | | Descriptor: | PROTEIN (3-ISOPROPYLMALATE DEHYDROGENASE) | | Authors: | Wallon, G, Kryger, G, Lovett, S.T, Oshima, T, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-05-17 | | Release date: | 1999-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structures of Escherichia coli and Salmonella typhimurium 3-isopropylmalate dehydrogenase and comparison with their thermophilic counterpart from Thermus thermophilus.
J.Mol.Biol., 266, 1997
|
|
1CNZ
 
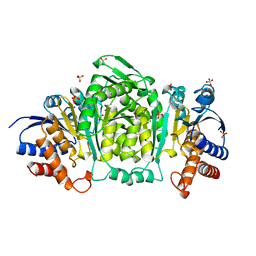 | | 3-ISOPROPYLMALATE DEHYDROGENASE (IPMDH) FROM SALMONELLA TYPHIMURIUM | | Descriptor: | MANGANESE (II) ION, PROTEIN (3-ISOPROPYLMALATE DEHYDROGENASE), SULFATE ION | | Authors: | Wallon, G, Kryger, G, Lovett, S.T, Oshima, T, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-05-24 | | Release date: | 1999-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structures of Eschericia Coli and Salmonella Typhimurium 3-
Isopropylmalate Dehydrogenase and Comparison with Their Thermophilic
Counterpart from Thermus Thermophilus
J.Mol.Biol., 266, 1997
|
|
1CM7
 
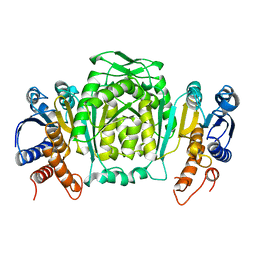 | | 3-ISOPROPYLMALATE DEHYDROGENASE FROM ESCHERICHIA COLI | | Descriptor: | PROTEIN (3-ISOPROPYLMALATE DEHYDROGENASE) | | Authors: | Wallon, G, Kryger, G, Lovett, S.T, Oshima, T, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-05-17 | | Release date: | 1999-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of Escherichia coli and Salmonella typhimurium 3-isopropylmalate dehydrogenase and comparison with their thermophilic counterpart from Thermus thermophilus.
J.Mol.Biol., 266, 1997
|
|
1CW7
 
 | |
1CW4
 
 | |
1CW1
 
 | |
1DPZ
 
 | | STRUCTURE OF MODIFIED 3-ISOPROPYLMALATE DEHYDROGENASE AT THE C-TERMINUS, HD711 | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Nurachman, Z, Akanuma, S, Sato, T, Oshima, T, Tanaka, N. | | Deposit date: | 1999-12-29 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of 3-isopropylmalate dehydrogenases with mutations at the C-terminus: crystallographic analyses of structure-stability relationships.
Protein Eng., 13, 2000
|
|
1DR8
 
 | | STRUCTURE OF MODIFIED 3-ISOPROPYLMALATE DEHYDROGENASE AT THE C-TERMINUS, HD177 | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Nurachman, Z, Akanuma, S, Sato, T, Oshima, T, Tanaka, N. | | Deposit date: | 2000-01-06 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of 3-isopropylmalate dehydrogenases with mutations at the C-terminus: crystallographic analyses of structure-stability relationships.
Protein Eng., 13, 2000
|
|
1DR0
 
 | | STRUCTURE OF MODIFIED 3-ISOPROPYLMALATE DEHYDROGENASE AT THE C-TERMINUS, HD708 | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Nurachman, Z, Akanuma, S, Sato, T, Oshima, T, Tanaka, N. | | Deposit date: | 2000-01-06 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of 3-isopropylmalate dehydrogenases with mutations at the C-terminus: crystallographic analyses of structure-stability relationships.
Protein Eng., 13, 2000
|
|
1GC8
 
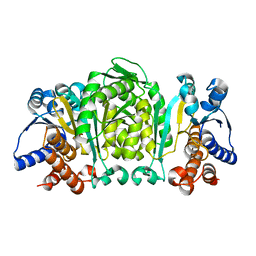 | | THE CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS 3-ISOPROPYLMALATE DEHYDROGENASE MUTATED AT 172TH FROM ALA TO PHE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Tanaka, N, Moriyama, H, Oshima, T. | | Deposit date: | 2000-07-27 | | Release date: | 2000-09-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, X-ray crystallography, molecular modelling and thermal stability studies of mutant enzymes at site 172 of 3-isopropylmalate dehydrogenase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GC9
 
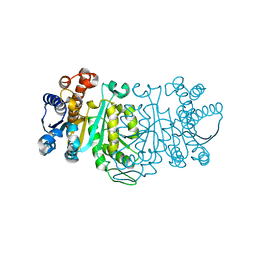 | | THE CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS 3-ISOPROPYLMALATE DEHYDROGENASE MUTATED AT 172TH FROM ALA TO GLY | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Tanaka, N, Moriyama, H, Oshima, T. | | Deposit date: | 2000-07-28 | | Release date: | 2000-09-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, X-ray crystallography, molecular modelling and thermal stability studies of mutant enzymes at site 172 of 3-isopropylmalate dehydrogenase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G2U
 
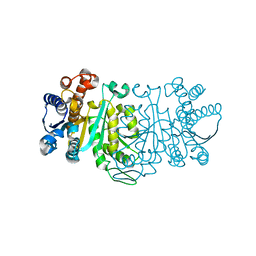 | | THE STRUCTURE OF THE MUTANT, A172V, OF 3-ISOPROPYLMALATE DEHYDROGENASE FROM THERMUS THERMOPHILUS HB8 : ITS THERMOSTABILITY AND STRUCTURE. | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Tanaka, N, Moriyama, H, Oshima, T. | | Deposit date: | 2000-10-21 | | Release date: | 2000-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, X-ray crystallography, molecular modelling and thermal stability studies of mutant enzymes at site 172 of 3-isopropylmalate dehydrogenase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HJ6
 
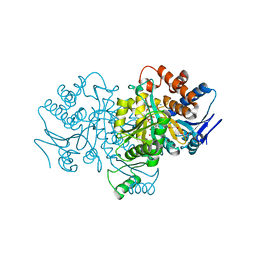 | | ISOCITRATE DEHYDROGENASE S113E MUTANT COMPLEXED WITH ISOPROPYLMALATE, NADP+ AND MAGNESIUM (FLASH-COOLED) | | Descriptor: | 3-ISOPROPYLMALIC ACID, GLYCEROL, ISOCITRATE DEHYDROGENASE, ... | | Authors: | Doyle, S.A, Beernink, P.T, Koshland Junior, D.E. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for a Change in Substrate Specificity: Crystal Structure of S113E Isocitrate Dehydrogenase in a Complex with Isopropylmalate, Mg2+ and Nadp
Biochemistry, 40, 2001
|
|
1HQS
 
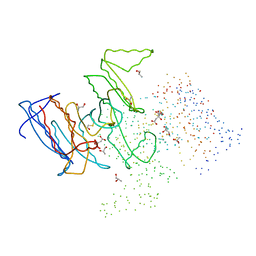 | | CRYSTAL STRUCTURE OF ISOCITRATE DEHYDROGENASE FROM BACILLUS SUBTILIS | | Descriptor: | CITRIC ACID, ISOCITRATE DEHYDROGENASE, R-1,2-PROPANEDIOL, ... | | Authors: | Singh, S.K, Matsuno, K, LaPorte, D.C, Banaszak, L.J. | | Deposit date: | 2000-12-19 | | Release date: | 2001-07-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of Bacillus subtilis isocitrate dehydrogenase at 1.55 A. Insights into the nature of substrate specificity exhibited by Escherichia coli isocitrate dehydrogenase kinase/phosphatase.
J.Biol.Chem., 276, 2001
|
|
1LWD
 
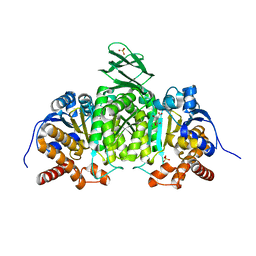 | |
1P8F
 
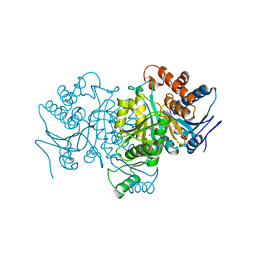 | |
1PB1
 
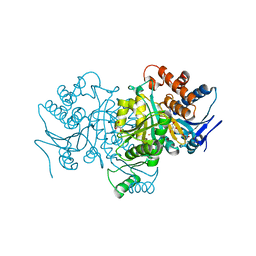 | |
1PB3
 
 | |
1T09
 
 | | Crystal structure of human cytosolic NADP(+)-dependent isocitrate dehydrogenase in complex NADP | | Descriptor: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Xu, X, Zhao, J, Peng, B, Huang, Q, Arnold, E, Ding, J. | | Deposit date: | 2004-04-08 | | Release date: | 2004-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity
J.Biol.Chem., 279, 2004
|
|
1T0L
 
 | | Crystal structure of human cytosolic NADP(+)-dependent isocitrate dehydrogenase in complex with NADP, isocitrate, and calcium(2+) | | Descriptor: | CALCIUM ION, ISOCITRIC ACID, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Xu, X, Zhao, J, Peng, B, Huang, Q, Arnold, E, Ding, J. | | Deposit date: | 2004-04-10 | | Release date: | 2004-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity
J.Biol.Chem., 279, 2004
|
|
