8IAP
 
 | | Respiratory complex Peripheral Arm of CI, focus-refined map of type I, Wild type mouse under thermoneutral temperature | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-02-09 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Respiratory complex Peripheral Arm of CI, focus-refined map of type I, Wild type mouse under thermoneutral temperature
To Be Published
|
|
8IBB
 
 | | Respiratory complex Membrane domain of CI, focus-refined of type IB, Wild type mouse under cold temperature | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-02-10 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Respiratory complex Membrane domain of CI, focus-refined of type IB, Wild type mouse under cold temperature
To Be Published
|
|
8IB6
 
 | | Respiratory complex Membrane domain of CI, focus-refined of type IA, Wild type mouse under cold temperature | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-02-09 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Respiratory complex Membrane domain of CI, focus-refined of type IA, Wild type mouse under cold temperature
To Be Published
|
|
9GAG
 
 | |
9FTI
 
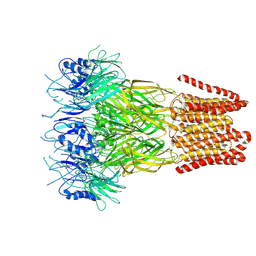 | | Open conformation of the pentameric ligand-gated ion channel, DeCLIC at pH 5 with 10mM EDTA | | Descriptor: | Neurotransmitter-gated ion-channel ligand-binding domain-containing protein | | Authors: | Rovsnik, U, Anden, O, Lycksell, M, Delarue, M, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural characterization of pH-modulated closed and open states
in a pentameric ligand-gated ion channel
To Be Published
|
|
9CBH
 
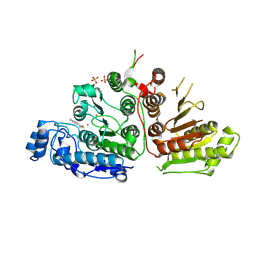 | |
9CF0
 
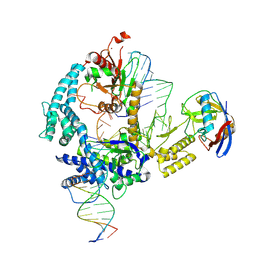 | | Parasitella parasitica Fanzor (PpFz) State 1 | | Descriptor: | DNA (5'-D(P*AP*CP*CP*CP*GP*GP*GP*AP*TP*AP*A)-3'), DNA (5'-D(P*TP*GP*TP*TP*TP*AP*TP*CP*CP*CP*GP*GP*GP*T)-3'), DNA/RNA (54-MER), ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 2024
|
|
9IZ6
 
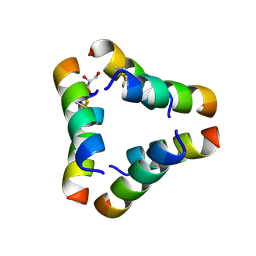 | |
9IMA
 
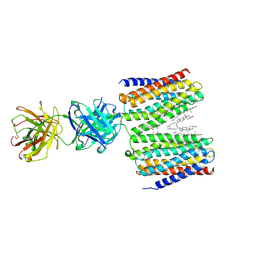 | | Cryo-EM structure for the GPRC5D complexed with Talquetamab Fab | | Descriptor: | CHOLESTEROL, G-protein coupled receptor family C group 5 member D, Talquetamab Fab (anti-GPRC5D) Heavy chain, ... | | Authors: | Jeong, J, Shin, J, Park, J, Cho, Y. | | Deposit date: | 2024-07-02 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural Basis for the Recognition of GPRC5D by Talquetamab, a Bispecific Antibody for Multiple Myeloma.
J.Mol.Biol., 436, 2024
|
|
9F1E
 
 | |
9GGQ
 
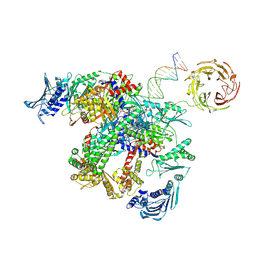 | | E.coli gyrase holocomplex with cleaved chirally wrapped 217 bp DNA fragment and moxifloxacin | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Ghilarov, D, Heddle, J.G, Pabis, M. | | Deposit date: | 2024-08-13 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of chiral wrap and T-segment capture by Escherichia coli DNA gyrase
Proceedings of the National Academy of Sciences USA, 2024
|
|
9IMP
 
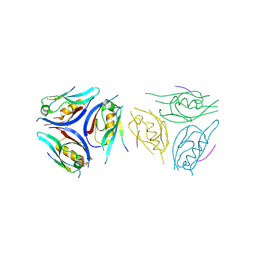 | | The complex of PDZ3 and PBM | | Descriptor: | INSC spindle orientation adaptor protein, Partitioning defective 3 homolog | | Authors: | Huang, S.J. | | Deposit date: | 2024-07-04 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | mInsc coordinates Par3 and NuMA condensates for assembly of the spindle orientation machinery in asymmetric cell division.
Int.J.Biol.Macromol., 279, 2024
|
|
9CET
 
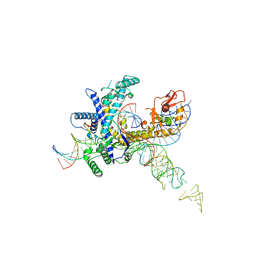 | | Guillardia theta Fanzor (GtFz) State 3 | | Descriptor: | DNA (28-MER), DNA (5'-D(P*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*TP*AP*AP*AP*GP*GP*CP*CP*CP*CP*GP*GP*G)-3'), Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 2024
|
|
9CER
 
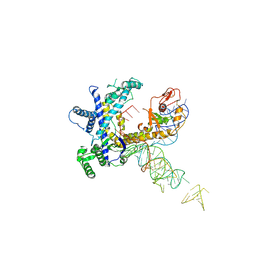 | | Guillardia theta Fanzor (GtFz) State 1 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein, Guillardia theta Fanzor1 chimera, RNA (142-MER) | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 2024
|
|
9FTJ
 
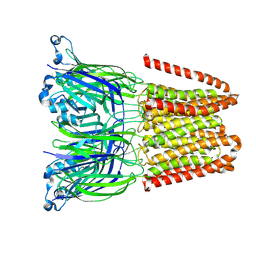 | | Closed and disordered conformation of the pentameric ligand-gated ion channel, DeCLIC at pH 5 with 10mM EDTA | | Descriptor: | Neurotransmitter-gated ion-channel ligand-binding domain-containing protein | | Authors: | Rovsnik, U, Anden, O, Lycksell, M, Delarue, M, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural characterization of pH-modulated closed and open states
in a pentameric ligand-gated ion channel
To Be Published
|
|
9CEZ
 
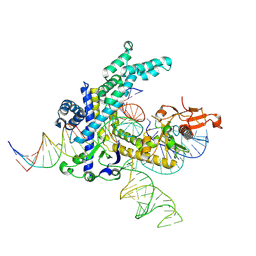 | | Spizellomyces punctatus Fanzor (SpuFz) State 6 | | Descriptor: | DNA (27-MER), DNA (5'-D(P*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 2024
|
|
9FCR
 
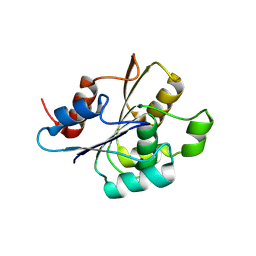 | |
9CLP
 
 | |
9D3G
 
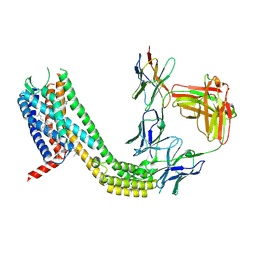 | | Cryo-EM structure of CCR6 bound by SQA1 and OXM1 | | Descriptor: | 1-(4-chlorophenyl)-N-{[(2R)-4-(2,3-dihydro-1H-inden-2-yl)-5-oxomorpholin-2-yl]methyl}cyclopropane-1-carboxamide, 4-[[3,4-bis(oxidanylidene)-2-[[(1~{R})-1-(4-propan-2-ylfuran-2-yl)propyl]amino]cyclobuten-1-yl]amino]-~{N},~{N}-dimethyl-3-oxidanyl-pyridine-2-carboxamide, CCR6, ... | | Authors: | Wasilko, D.J, Wu, H. | | Deposit date: | 2024-08-10 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for CCR6 modulation by allosteric antagonists.
Nat Commun, 15, 2024
|
|
9BQJ
 
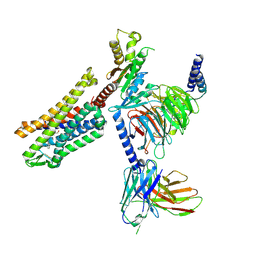 | | RO76 bound muOR-Gi1-scFv16 complex structure | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, H, Majumdar, S, Kobilka, B.K. | | Deposit date: | 2024-05-10 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Signaling Modulation Mediated by Ligand Water Interactions with the Sodium Site at mu OR.
Acs Cent.Sci., 10, 2024
|
|
9F1M
 
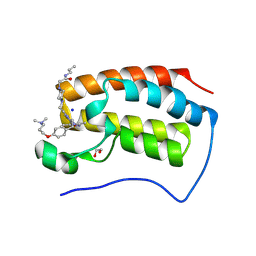 | | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 45 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-[7-[2-[4-[2-[2-[2-[4-[3-(dimethylamino)propoxy]phenyl]ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzimidazol-1-yl]ethyl]piperazin-1-yl]ethanoylamino]heptanoylamino]-3,3-dimethyl-butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, ... | | Authors: | Balourdas, D.I, Edmonds, A.K, Marsh, G.P, Maple, H.J, Spencer, J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.90001261 Å) | | Cite: | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 45
to be published
|
|
9C1A
 
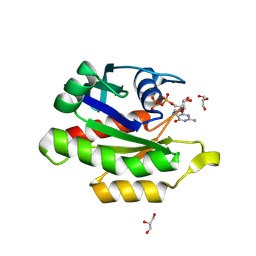 | | Crystal structure of GDP-bound human M-RAS protein in crystal form I | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bester, S.M, Abrahamsen, R, Samora, L.R, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-05-28 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of the GDP-bound human M-RAS protein in two crystal forms.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
9F1N
 
 | | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 46 | | Descriptor: | (2S,4R)-1-[(2S)-2-[9-[2-[4-[2-[2-[2-[4-[3-(dimethylamino)propoxy]phenyl]ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzimidazol-1-yl]ethyl]piperazin-1-yl]ethanoylamino]nonanoylamino]-3,3-dimethyl-butanoyl]-N-[[4-(4-methyl-4H-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Balourdas, D.I, Edmonds, A.K, Marsh, G.P, Maple, H.J, Spencer, J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.71000016 Å) | | Cite: | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 46
to be published
|
|
9GG1
 
 | | P301T type I tau filaments from human brain | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Schweighauser, M, Shi, Y, Murzin, A.G, Garringer, H.J, Vidal, R, Murrell, J.R, Erro, M.E, Seelaar, H, Ferrer, I, van Swieten, J.C, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2024-08-12 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Novel tau filament folds in individuals with MAPT mutations P301L and P301T.
Biorxiv, 2024
|
|
9FLB
 
 | |
