9JBE
 
 | | Cryo-EM structure of the human LYCHOS in complex with cholesterol and cholesteryl hemisuccinate in the contracted state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL, ... | | Authors: | Yu, S, Liang, L. | | Deposit date: | 2024-08-27 | | Release date: | 2025-07-16 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural insights into cholesterol sensing by the LYCHOS-mTORC1 pathway.
Nat Commun, 16, 2025
|
|
9JBF
 
 | | Cryo-EM structure of the human LYCHOS Y57A mutant in complex with cholesteryl hemisuccinate in the contracted state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL, ... | | Authors: | Yu, S, Liang, L. | | Deposit date: | 2024-08-27 | | Release date: | 2025-07-16 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural insights into cholesterol sensing by the LYCHOS-mTORC1 pathway.
Nat Commun, 16, 2025
|
|
9JBG
 
 | | Cryo-EM structure of the human LYCHOS in complex with lipids in the expanded state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Lysosomal cholesterol signaling protein, ... | | Authors: | Yu, S, Liang, L. | | Deposit date: | 2024-08-27 | | Release date: | 2025-07-16 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural insights into cholesterol sensing by the LYCHOS-mTORC1 pathway.
Nat Commun, 16, 2025
|
|
9JBH
 
 | | Cryo-EM structure of the human LYCHOS PLD homodimer | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, Lysosomal cholesterol signaling protein | | Authors: | Yu, S, Liang, L. | | Deposit date: | 2024-08-27 | | Release date: | 2025-07-16 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural insights into cholesterol sensing by the LYCHOS-mTORC1 pathway.
Nat Commun, 16, 2025
|
|
9JBI
 
 | |
9JBJ
 
 | |
9JQG
 
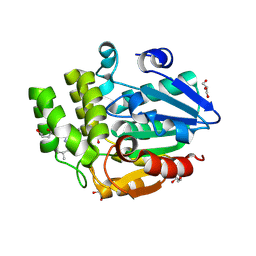 | | Crystal structure of rice DWARF14 in complex with Cyclo(L-Leu-L-Pro) | | Descriptor: | (3~{S},8~{a}~{S})-3-(2-methylpropyl)-2,3,6,7,8,8~{a}-hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, 1,3-PROPANDIOL, 1,4-BUTANEDIOL, ... | | Authors: | Wang, Q.X, Wang, M.Y, Gao, S, Wang, B, Bai, Y. | | Deposit date: | 2024-09-27 | | Release date: | 2025-05-21 | | Last modified: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Root microbiota regulates tiller number in rice.
Cell, 188, 2025
|
|
9JWY
 
 | | Crystal structure of PstS from Candidatus Pelagibacter sp. HTCC7211 in complex with phosphate | | Descriptor: | PHOSPHATE ION, Phosphate ABC transporter, periplasmic phosphate-binding protein | | Authors: | Zhu, W.J, Wang, P, Cao, H.Y, Zhang, Y.Z. | | Deposit date: | 2024-10-11 | | Release date: | 2025-07-30 | | Last modified: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and molecular basis for phosphate recognition by SAR11 bacteria.
Mbio, 2025
|
|
9KHS
 
 | |
9KHT
 
 | |
9L0D
 
 | | Cryo-EM structure of the human MON1A/CCZ1/C18orf8 complex | | Descriptor: | Ras-related protein Rab-7a, Regulator of MON1-CCZ1 complex, Vacuolar fusion protein CCZ1 homolog B, ... | | Authors: | Tang, Y, Han, Y, Zhang, Y, Pan, L. | | Deposit date: | 2024-12-12 | | Release date: | 2025-03-19 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Mechanistic insights into the GEF activity of the human MON1A/CCZ1/C18orf8 complex.
Protein Cell, 16, 2025
|
|
9L7L
 
 | |
9LE4
 
 | | Crystal structure of the MIT-CD complex of STAMBP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, STAM-binding protein, ZINC ION | | Authors: | Chen, Z, Ding, J. | | Deposit date: | 2025-01-07 | | Release date: | 2025-06-11 | | Last modified: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The MIT domain of STAMBP autoinhibits its deubiquitination activity.
Structure, 33, 2025
|
|
9LVR
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, 6-(1,3-dihydroisoindol-2-yl)-3-(5-methylpyridin-3-yl)-1-[[3,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Unoh, Y, Hirai, K, Uehara, S, Kawashima, S, Nobori, H, Sato, J, Shibayama, H, Hori, A, Nakahara, K, Kurahashi, K, Takamatsu, M, Yamamoto, S, Zhang, O, Tanimura, M, Dodo, R, Maruyama, Y, Sawa, H, Watari, R, Miyano, T, Kato, T, Sato, T, Tachibana, Y. | | Deposit date: | 2025-02-12 | | Release date: | 2025-05-14 | | Last modified: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of the Clinical Candidate S-892216 : A Second-Generation of SARS-CoV-2 3CL Protease Inhibitor for Treating COVID-19.
J.Med.Chem., 2025
|
|
9LVT
 
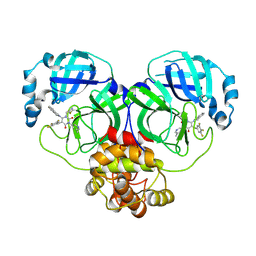 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 4 | | Descriptor: | 1-(2-azanylideneethyl)-6-(1,3-dihydroisoindol-2-yl)-3-(5-methylpyridin-3-yl)-5-[[3,4,5-tris(fluoranyl)phenyl]methyl]pyrimidine-2,4-dione, 3C-like proteinase | | Authors: | Unoh, Y, Hirai, K, Uehara, S, Kawashima, S, Nobori, H, Sato, J, Shibayama, H, Hori, A, Nakahara, K, Kurahashi, K, Takamatsu, M, Yamamoto, S, Zhang, O, Tanimura, M, Dodo, R, Maruyama, Y, Sawa, H, Watari, R, Miyano, T, Kato, T, Sato, T, Tachibana, Y. | | Deposit date: | 2025-02-12 | | Release date: | 2025-05-14 | | Last modified: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of the Clinical Candidate S-892216 : A Second-Generation of SARS-CoV-2 3CL Protease Inhibitor for Treating COVID-19.
J.Med.Chem., 2025
|
|
9LVV
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 17 (S-892216) | | Descriptor: | 1-(2-azanylideneethyl)-6-[6,6-bis(fluoranyl)-2-azaspiro[3.3]heptan-2-yl]-5-(3-chloranyl-4-fluoranyl-phenyl)-3-(5-chloranylpyridin-3-yl)pyrimidine-2,4-dione, 3C-like proteinase | | Authors: | Unoh, Y, Hirai, K, Uehara, S, Kawashima, S, Nobori, H, Sato, J, Shibayama, H, Hori, A, Nakahara, K, Kurahashi, K, Takamatsu, M, Yamamoto, S, Zhang, O, Tanimura, M, Dodo, R, Maruyama, Y, Sawa, H, Watari, R, Miyano, T, Kato, T, Sato, T, Tachibana, Y. | | Deposit date: | 2025-02-12 | | Release date: | 2025-05-14 | | Last modified: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of the Clinical Candidate S-892216 : A Second-Generation of SARS-CoV-2 3CL Protease Inhibitor for Treating COVID-19.
J.Med.Chem., 2025
|
|
9LXL
 
 | | Crystal structure of GH29 family alpha-L-fucosidase from Fusarium proliferatum LE1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Korban, S.A, Borshchevskiy, V.I, Pospelov, V.A, Bobrov, K.S, Ivanova, D.N, Kulminskaya, A.A. | | Deposit date: | 2025-02-18 | | Release date: | 2025-03-19 | | Last modified: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Structure and biochemical characterization of GH29 family alpha-l-fucosidase from Fusarium proliferatum LE1.
Biochem.Biophys.Res.Commun., 779, 2025
|
|
9M7O
 
 | |
9MQF
 
 | |
9MYA
 
 | |
9MYB
 
 | |
9N1A
 
 | | crystal structure of diferric HrmI from Streptomyces griseoflavus | | Descriptor: | FE (III) ION, GLYCEROL, HrmI | | Authors: | Skirboll, S.S, Phan, H.N, Makris, T.M, Swartz, P.D. | | Deposit date: | 2025-01-24 | | Release date: | 2025-08-20 | | Last modified: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The Heme Oxygenase-Like Diiron Enzyme HrmI Reveals Altered Regulatory Mechanisms for Dioxygen Activation and Substrate N-Oxygenation.
J.Am.Chem.Soc., 147, 2025
|
|
9N1E
 
 | | Crystal structure of N-oxygenase HrmI with the diferric cofactor partially loaded | | Descriptor: | FE (III) ION, GLYCEROL, HrmI, ... | | Authors: | Skirboll, S.S, Phan, H.N, Swartz, P.D, Makris, T.M. | | Deposit date: | 2025-01-25 | | Release date: | 2025-08-20 | | Last modified: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Heme Oxygenase-Like Diiron Enzyme HrmI Reveals Altered Regulatory Mechanisms for Dioxygen Activation and Substrate N-Oxygenation.
J.Am.Chem.Soc., 147, 2025
|
|
9N1X
 
 | | Crystal Structure of N-oxygenase HrmI with the diferrous cofactor | | Descriptor: | FE (II) ION, GLYCEROL, HrmI, ... | | Authors: | Skirboll, S.S, Phan, H.N, Swartz, P.D, Makris, T.M. | | Deposit date: | 2025-01-27 | | Release date: | 2025-08-20 | | Last modified: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The Heme Oxygenase-Like Diiron Enzyme HrmI Reveals Altered Regulatory Mechanisms for Dioxygen Activation and Substrate N-Oxygenation.
J.Am.Chem.Soc., 147, 2025
|
|
9N2A
 
 | | Crystal structure of N-oxygenase HrmI with the diferrous cofactor and substrate bound | | Descriptor: | FE (II) ION, GLYCEROL, HrmI, ... | | Authors: | Skirboll, S.S, Phan, H.N, Swartz, P.D, Makris, T.M. | | Deposit date: | 2025-01-28 | | Release date: | 2025-08-20 | | Last modified: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The Heme Oxygenase-Like Diiron Enzyme HrmI Reveals Altered Regulatory Mechanisms for Dioxygen Activation and Substrate N-Oxygenation.
J.Am.Chem.Soc., 147, 2025
|
|
