9HMB
 
 | | Crystal structure of GH139 glycoside hydrolase from Verrucomicrobium sp. in the hexagonal space group P6522 | | Descriptor: | DUF5703 domain-containing protein, GLYCEROL | | Authors: | Moraleda-Montoya, A, Garcia-Alija, M, Trastoy, B, Guerin, M. | | Deposit date: | 2024-12-08 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Understanding the substrate recognition and catalytic mechanism of 2-O-methyl fucosidases from glycoside hydrolase family 139.
J.Biol.Chem., 301, 2025
|
|
9HQ6
 
 | | Structural insights in the HuHf@gold-monocarbene adduct: aurophilicity revealed in a biological context | | Descriptor: | 1-butyl-3-methyl-1H-imidazol-3-ium, Ferritin heavy chain, GOLD ION | | Authors: | Cosottini, L, Giachetti, A, Guerri, A, Martinez-Castillo, A, Geri, A, Zineddu, S, Abrescia, N.G.A, Messori, L, Turano, P, Rosato, A. | | Deposit date: | 2024-12-16 | | Release date: | 2025-05-07 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (1.51 Å) | | Cite: | Structural Insight Into a Human H Ferritin@Gold-Monocarbene Adduct: Aurophilicity Revealed in a Biological Context.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9HRT
 
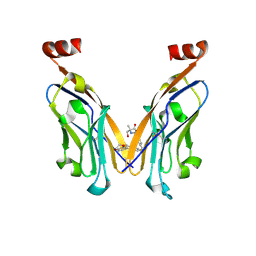 | | Structure of human PD-L1 in complex with clinically evaluated inhibitor | | Descriptor: | (2~{R})-2-[[3-[(~{E})-2-[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]ethenyl]-4-(trifluoromethyl)phenyl]methylamino]-2-methyl-3-oxidanyl-propanoic acid, Programmed cell death 1 ligand 1 | | Authors: | Slota, A, Golebiowska-Mendroch, K, Plewka, J, Magiera-Mularz, K. | | Deposit date: | 2024-12-18 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of Clinically Evaluated Small-Molecule Inhibitors of PD-L1 for Immunotherapy.
Acs Med.Chem.Lett., 16, 2025
|
|
9HYQ
 
 | |
9HZ5
 
 | | Pre-clinical characterization of novel multi-client inhibitors of Sec61 with broad anti-tumor activity | | Descriptor: | 4-{(3S)-9-(cyclohexylmethyl)-5-[(3R,5R)-4-(3-fluoro-5-methoxyphenyl)-3,5-dimethylpiperazine-1-sulfonyl]-3-methyl-1,5,9-triazacyclododecane-1-sulfonyl}-N,N-dimethylaniline, Protein transport protein Sec61 subunit alpha, Protein transport protein Sec61 subunit beta, ... | | Authors: | Shahid, R, Paavilainen, V.O. | | Deposit date: | 2025-01-13 | | Release date: | 2025-07-16 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Preclinical characterization of novel multi-client inhibitors of Sec61 with broad antitumor activity.
J.Pharmacol.Exp.Ther., 392, 2025
|
|
9I0U
 
 | | Structure of human PD-L1 in complex with clinically evaluated inhibitor | | Descriptor: | (5~{S})-5-[[[5-[2-chloranyl-3-[2-chloranyl-3-[6-methoxy-5-[[[(2~{S})-5-oxidanylidenepyrrolidin-2-yl]methylamino]methyl]pyrazin-2-yl]phenyl]phenyl]-3-methoxy-pyrazin-2-yl]methylamino]methyl]pyrrolidin-2-one, GLYCEROL, Programmed cell death 1 ligand 1 | | Authors: | Golebiowska-Mendroch, K, Slota, A, Plewka, J, Magiera-Mularz, K. | | Deposit date: | 2025-01-15 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Characterization of Clinically Evaluated Small-Molecule Inhibitors of PD-L1 for Immunotherapy.
Acs Med.Chem.Lett., 16, 2025
|
|
9I0W
 
 | | Structure of human PD-L1 in complex with clinically evaluated inhibitor | | Descriptor: | (3~{R})-1-[[7-(iminomethyl)-2-[2-methyl-3-[2-methyl-3-[[3-[[(3~{R})-3-oxidanylpyrrolidin-1-yl]methyl]-1,7-naphthyridin-8-yl]amino]phenyl]phenyl]-1,3-benzoxazol-5-yl]methyl]pyrrolidine-3-carboxylic acid, Programmed cell death 1 ligand 1 | | Authors: | Plewka, J, Golebiowska-Mendroch, K, Slota, A, Magiera-Mularz, K. | | Deposit date: | 2025-01-15 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of Clinically Evaluated Small-Molecule Inhibitors of PD-L1 for Immunotherapy.
Acs Med.Chem.Lett., 16, 2025
|
|
9I6B
 
 | | CryoEM structure of the Chaetomium thermophilum TOM core complex at 2.7 angstrom resolution (pALDH treated) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Agip, A.N.A, Ornelas, P, Yang, T.J, Ermanno, U, Haeder, S, McDowell, M.A, Kuehlbrandt, W. | | Deposit date: | 2025-01-29 | | Release date: | 2025-07-09 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of Chaetomium thermophilum TOM complexes with bound preproteins.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9I7P
 
 | | CryoEM structure of the Chaetomium thermophilum TOM core complex at 3.2 angstrom resolution | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Agip, A.N.A, Ornelas, P, Yang, T.J, Ermanno, U, Haeder, S, McDowell, M.A, Kuehlbrandt, W. | | Deposit date: | 2025-01-31 | | Release date: | 2025-07-09 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of Chaetomium thermophilum TOM complexes with bound preproteins.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9I7S
 
 | | CryoEM structure of the Chaetomium thermophilum TOM holo complex at 3.2 angstrom resolution (pALDH treated) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Agip, A.N.A, Ornelas, P, Yang, T.J, Ermanno, U, Haeder, S, McDowell, M.A, Kuehlbrandt, W. | | Deposit date: | 2025-02-01 | | Release date: | 2025-07-09 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of Chaetomium thermophilum TOM complexes with bound preproteins.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9I7T
 
 | | CryoEM structure of the Chaetomium thermophilum TOM holo complex at 3.8 angstrom resolution | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Agip, A.N.A, Ornelas, P, Yang, T.J, Ermanno, U, Haeder, S, McDowell, M.A, Kuehlbrandt, W. | | Deposit date: | 2025-02-01 | | Release date: | 2025-07-09 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of Chaetomium thermophilum TOM complexes with bound preproteins.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9IGQ
 
 | | Crystal structure of PPK2 class III from Erysipelotrichaceae bacterium in complex with AppCH2p and polyphosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BENZOIC ACID, GLYCEROL, ... | | Authors: | Rasche, R, Lawrence-Doerner, A.-M, Cornelissen, N.V, Kuemmel, D. | | Deposit date: | 2025-02-20 | | Release date: | 2025-07-23 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-guided engineering of a polyphosphate kinase 2 class III from an Erysipelotrichaceae bacterium to produce base-modified purine nucleotides.
Rsc Chem Biol, 2025
|
|
9IGR
 
 | | Crystal structure of PPK2 class III from Erysipelotrichaceae bacterium in complex with polyphosphate | | Descriptor: | BENZOIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Rasche, R, Lawrence-Doerner, A.-M, Cornelissen, N.V, Kuemmel, D. | | Deposit date: | 2025-02-20 | | Release date: | 2025-07-23 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | Structure-guided engineering of a polyphosphate kinase 2 class III from an Erysipelotrichaceae bacterium to produce base-modified purine nucleotides.
Rsc Chem Biol, 2025
|
|
9IIU
 
 | | Cryo-EM structure of an TEF30-associated intermediate PSII core complex from Chlamydomonas reinhardtii | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Wang, Y, Wang, C, Li, A, Liu, Z. | | Deposit date: | 2024-06-21 | | Release date: | 2025-05-14 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Roles of multiple TEF30-associated intermediate complexes in the repair and reassembly of photosystem II in Chlamydomonas reinhardtii.
Nat.Plants, 11, 2025
|
|
9IMN
 
 | | Cryo-EM structure of a TEF30-associated intermediate PSII core dimer complex, type I, from Chlamydomonas reinhardtii | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Wang, Y, Wang, C, Li, A, Liu, Z. | | Deposit date: | 2024-07-03 | | Release date: | 2025-05-14 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Roles of multiple TEF30-associated intermediate complexes in the repair and reassembly of photosystem II in Chlamydomonas reinhardtii.
Nat.Plants, 11, 2025
|
|
9IOL
 
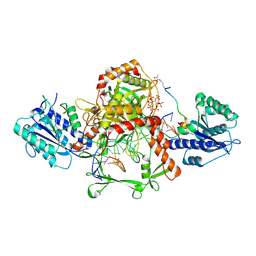 | | Cryo-EM structure of the complex of DNA, Ku70/80, and laXLF. | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, DNA (5'-D(P*AP*GP*GP*TP*CP*GP*AP*CP*GP*AP*AP*TP*CP*GP*GP*CP*AP*GP*CP*G)-3'), DNA (5'-D(P*CP*GP*CP*TP*GP*CP*CP*GP*AP*TP*TP*CP*GP*TP*CP*GP*AP*CP*CP*T)-3'), ... | | Authors: | Liang, S. | | Deposit date: | 2024-07-09 | | Release date: | 2025-07-09 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Lactylation of XLF promotes non-homologous end-joining repair and chemoresistance in cancer.
Mol.Cell, 85, 2025
|
|
9IOX
 
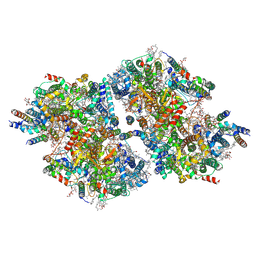 | | Cryo-EM structure of a TEF30-associated intermediate PSII core dimer complex, type II, from Chlamydomonas reinhardtii | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Wang, Y, Wang, C, Li, A, Liu, Z. | | Deposit date: | 2024-07-09 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Roles of multiple TEF30-associated intermediate complexes in the repair and reassembly of photosystem II in Chlamydomonas reinhardtii.
Nat.Plants, 11, 2025
|
|
9IP7
 
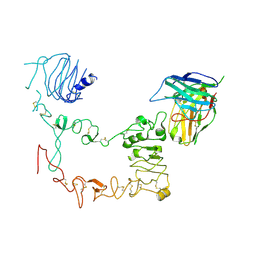 | | Local refinement structure of sEGFR and 528 Fv (from HL-type bispecific diabody Ex3) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 528 Fv from HL-type bispecific diabody Ex3, ... | | Authors: | Sato, K, Uehara, S, Tsugita, A, Matsui, T, Asano, R, Makabe, K, Yokoyama, T, Tanaka, Y. | | Deposit date: | 2024-07-10 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Bispecific antibody-antigen complex structures reveal activity enhancement by domain rearrangement.
Cell Rep, 2025
|
|
9IP8
 
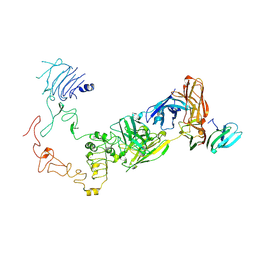 | | Poly-alanine model for HL-type bispecific diabody Ex3 composed of 528 and OKT3 Fvs in ternary complex with sEGFR and CD3gamma-epsilon (closed conformation) | | Descriptor: | Epidermal growth factor receptor, HL-type bispecific diabody Ex3, T-cell surface glycoprotein CD3 gamma chain,T-cell surface glycoprotein CD3 epsilon chain | | Authors: | Sato, K, Uehara, S, Tsugita, A, Matsui, T, Asano, R, Makabe, K, Yokoyama, T, Tanaka, Y. | | Deposit date: | 2024-07-10 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Bispecific antibody-antigen complex structures reveal activity enhancement by domain rearrangement.
Cell Rep, 2025
|
|
9IP9
 
 | | Poly-alanine model for HL-type bispecific diabody Ex3 composed of 528 and OKT3 Fvs in ternary complex with sEGFR and CD3gamma-epsilon (middle conformation) | | Descriptor: | Epidermal growth factor receptor, HL-type bispecific diabody Ex3, T-cell surface glycoprotein CD3 gamma chain,T-cell surface glycoprotein CD3 epsilon chain | | Authors: | Sato, K, Uehara, S, Tsugita, A, Matsui, T, Asano, R, Makabe, K, Yokoyama, T, Tanaka, Y. | | Deposit date: | 2024-07-10 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Bispecific antibody-antigen complex structures reveal activity enhancement by domain rearrangement.
Cell Rep, 2025
|
|
9IPA
 
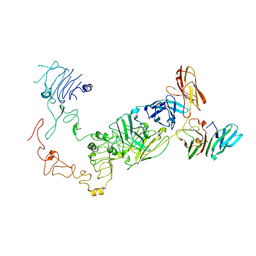 | | Poly-alanine model for HL-type bispecific diabody Ex3 composed of 528 and OKT3 Fvs in ternary complex with sEGFR and CD3gamma-epsilon (open conformation) | | Descriptor: | Epidermal growth factor receptor, HL-type bispecific diabody Ex3, T-cell surface glycoprotein CD3 gamma chain,T-cell surface glycoprotein CD3 epsilon chain | | Authors: | Sato, K, Uehara, S, Tsugita, A, Matsui, T, Asano, R, Makabe, K, Yokoyama, T, Tanaka, Y. | | Deposit date: | 2024-07-10 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Bispecific antibody-antigen complex structures reveal activity enhancement by domain rearrangement.
Cell Rep, 2025
|
|
9IPB
 
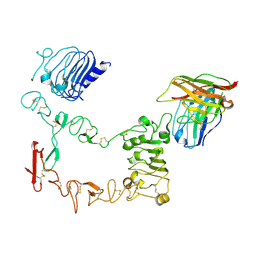 | | Local refinement structure of sEGFR and 528 Fv (from LH-type bispecific diabody Ex3) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 528 Fv from LH-type bispecific diabody Ex3, ... | | Authors: | Sato, K, Uehara, S, Tsugita, A, Matsui, T, Asano, R, Makabe, K, Yokoyama, T, Tanaka, Y. | | Deposit date: | 2024-07-10 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Bispecific antibody-antigen complex structures reveal activity enhancement by domain rearrangement.
Cell Rep, 2025
|
|
9IPC
 
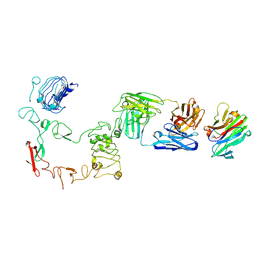 | | Poly-alanine model for LH-type bispecific diabody Ex3 composed of 528 and OKT3 Fvs in ternary complex with sEGFR and CD3gamma-epsilon (closed conformation) | | Descriptor: | Epidermal growth factor receptor, LH-type bispecific diabody Ex3, T-cell surface glycoprotein CD3 gamma chain,T-cell surface glycoprotein CD3 epsilon chain | | Authors: | Sato, K, Uehara, S, Tsugita, A, Matsui, T, Asano, R, Makabe, K, Yokoyama, T, Tanaka, Y. | | Deposit date: | 2024-07-10 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Bispecific antibody-antigen complex structures reveal activity enhancement by domain rearrangement.
Cell Rep, 2025
|
|
9IPD
 
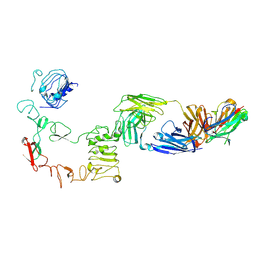 | | Poly-alanine model for LH-type bispecific diabody Ex3 composed of 528 and OKT3 Fvs in ternary complex with sEGFR and CD3gamma-epsilon (middle conformation) | | Descriptor: | Epidermal growth factor receptor, LH-type bispecific diabody Ex3, T-cell surface glycoprotein CD3 gamma chain,T-cell surface glycoprotein CD3 epsilon chain | | Authors: | Sato, K, Uehara, S, Tsugita, A, Matsui, T, Asano, R, Makabe, K, Yokoyama, T, Tanaka, Y. | | Deposit date: | 2024-07-10 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Bispecific antibody-antigen complex structures reveal activity enhancement by domain rearrangement.
Cell Rep, 2025
|
|
9IPE
 
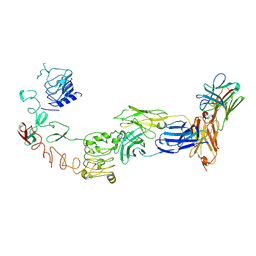 | | Poly-alanine model for LH-type bispecific diabody Ex3 composed of 528 and OKT3 Fvs in ternary complex with sEGFR and CD3gamma-epsilon (open conformation) | | Descriptor: | Epidermal growth factor receptor, LH-type bispecific diabody Ex3, T-cell surface glycoprotein CD3 gamma chain,T-cell surface glycoprotein CD3 epsilon chain | | Authors: | Sato, K, Uehara, S, Tsugita, A, Matsui, T, Asano, R, Makabe, K, Yokoyama, T, Tanaka, Y. | | Deposit date: | 2024-07-10 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Bispecific antibody-antigen complex structures reveal activity enhancement by domain rearrangement.
Cell Rep, 2025
|
|
