5TQW
 
 | | CryoEM reconstruction of human IKK1, open conformation 1 | | Descriptor: | Inhibitor of nuclear factor kappa-B kinase subunit alpha | | Authors: | Lyumkis, D, Ghosh, G, Polley, S, Biswas, T, Huang, D, Passos, D.O. | | Deposit date: | 2016-10-24 | | Release date: | 2016-11-09 | | Last modified: | 2026-02-25 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural Basis for the Activation of IKK1/ alpha.
Cell Rep, 17, 2016
|
|
5TQX
 
 | | CryoEM reconstruction of human IKK1, intermediate conformation 2 | | Descriptor: | Inhibitor of nuclear factor kappa-B kinase subunit alpha | | Authors: | Lyumkis, D, Ghosh, G, Polley, S, Biswas, T, Huang, D, Passos, D.O. | | Deposit date: | 2016-10-24 | | Release date: | 2016-11-09 | | Last modified: | 2026-02-25 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structural Basis for the Activation of IKK1/ alpha.
Cell Rep, 17, 2016
|
|
5TQY
 
 | | CryoEM reconstruction of human IKK1, closed conformation 3 | | Descriptor: | Inhibitor of nuclear factor kappa-B kinase subunit alpha | | Authors: | Lyumkis, D, Ghosh, G, Polley, S, Biswas, T, Huang, D, Passos, D.O. | | Deposit date: | 2016-10-24 | | Release date: | 2016-11-09 | | Last modified: | 2026-02-25 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structural Basis for the Activation of IKK1/ alpha.
Cell Rep, 17, 2016
|
|
5TVL
 
 | | Crystal structure of foldase protein PrsA from Streptococcus pneumoniae str. Canada MDR_19A | | Descriptor: | CHLORIDE ION, Foldase protein PrsA, GLYCEROL, ... | | Authors: | Borek, D, Yim, V, Kudritska, M, Wawrzak, Z, Stogios, P.J, Otwinowski, Z, Savchenko, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-09 | | Release date: | 2016-11-23 | | Last modified: | 2026-02-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural analysis of extracellular ATP-independent chaperones of streptococcal species and protein substrate interactions.
Msphere, 10, 2025
|
|
5VGC
 
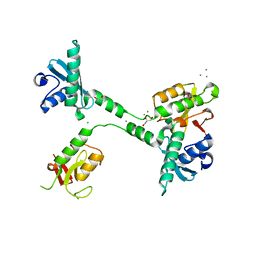 | | Crystal structure of the NleG5-1 effector (C200A) from Escherichia coli O157:H7 str. Sakai | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Borek, D, Valleau, D, Skarina, T, Jobin, M.C, Wawrzak, Z, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-18 | | Last modified: | 2026-02-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional diversification of the NleG effector family in enterohemorrhagic Escherichia coli.
Proc.Natl.Acad.Sci.USA, 115, 2018
|
|
6K4I
 
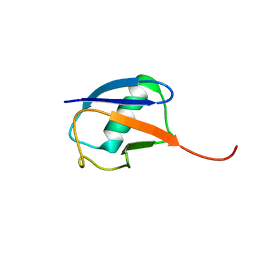 | | The partially disordered conformation of ubiquitin (Q41N variant) | | Descriptor: | ubiquitin | | Authors: | Wakamoto, T, Ikeya, T, Kitazawa, S, Baxter, N.J, Williamson, M.P, Kitahara, R. | | Deposit date: | 2019-05-24 | | Release date: | 2019-10-30 | | Last modified: | 2026-02-25 | | Method: | SOLUTION NMR | | Cite: | Paramagnetic relaxation enhancement-assisted structural characterization of a partially disordered conformation of ubiquitin.
Protein Sci., 28, 2019
|
|
6OKZ
 
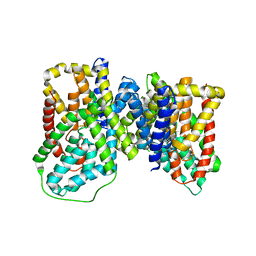 | | Structure of VcINDY bound to Fumarate | | Descriptor: | FUMARIC ACID, SODIUM ION, Transporter, ... | | Authors: | Sauer, D.B, Marden, J, Wang, D.N. | | Deposit date: | 2019-04-15 | | Release date: | 2020-10-28 | | Last modified: | 2026-02-25 | | Method: | X-RAY DIFFRACTION (3.292 Å) | | Cite: | Elevator mechanism dynamics in a sodium-coupled dicarboxylate transporter.
Proc.Natl.Acad.Sci.USA, 123, 2026
|
|
6OL0
 
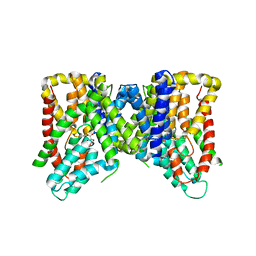 | | Structure of VcINDY bound to Malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, SODIUM ION, Transporter, ... | | Authors: | Sauer, D.B, Marden, J.J, Wang, D.N. | | Deposit date: | 2019-04-15 | | Release date: | 2020-10-28 | | Last modified: | 2026-02-25 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Elevator mechanism dynamics in a sodium-coupled dicarboxylate transporter.
Proc.Natl.Acad.Sci.USA, 123, 2026
|
|
6OL1
 
 | | Structure of VcINDY in complex with Succinate | | Descriptor: | SODIUM ION, SUCCINIC ACID, Transporter, ... | | Authors: | Sauer, D.B, Marden, J.J, Wang, D.N. | | Deposit date: | 2019-04-15 | | Release date: | 2020-10-28 | | Last modified: | 2026-02-25 | | Method: | X-RAY DIFFRACTION (3.088 Å) | | Cite: | Elevator mechanism dynamics in a sodium-coupled dicarboxylate transporter.
Proc.Natl.Acad.Sci.USA, 123, 2026
|
|
6SG2
 
 | | FeFe Hydrogenase from Nitratidesulfovibrio vulgaris in Hinact state | | Descriptor: | HydB, IRON/SULFUR CLUSTER, Periplasmic [Fe] hydrogenase large subunit, ... | | Authors: | Galle, L.M, Span, I. | | Deposit date: | 2019-08-02 | | Release date: | 2020-07-08 | | Last modified: | 2026-02-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Caught in the H inact : Crystal Structure and Spectroscopy Reveal a Sulfur Bound to the Active Site of an O 2 -stable State of [FeFe] Hydrogenase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6UQF
 
 | | Human HCN1 channel in a hyperpolarized conformation | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, MERCURY (II) ION, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Lee, C.-H, MacKinnon, R. | | Deposit date: | 2019-10-19 | | Release date: | 2019-12-11 | | Last modified: | 2026-02-25 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Voltage Sensor Movements during Hyperpolarization in the HCN Channel.
Cell, 179, 2019
|
|
7QVD
 
 | |
7UUJ
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with gentamicin | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, ... | | Authors: | Stogios, P.J, Evdokimova, E, Osipiuk, J, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2023-04-19 | | Last modified: | 2026-02-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance
Nat.Chem.Biol., 20, 2024
|
|
7UUK
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with tobramycin | | Descriptor: | Aminocyclitol acetyltransferase ApmA, CHLORIDE ION, TOBRAMYCIN | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2023-04-19 | | Last modified: | 2026-02-25 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance
Nat.Chem.Biol., 20, 2024
|
|
7UUL
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with kanamycin B and coenzyme A | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2026-02-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance
Nat.Chem.Biol., 20, 2024
|
|
7UUM
 
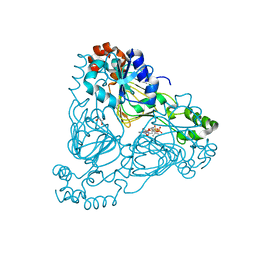 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with paromomycin and coenzyme A | | Descriptor: | Aminocyclitol acetyltransferase ApmA, COENZYME A, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Osipiuk, J, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2026-02-25 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance
Nat.Chem.Biol., 20, 2024
|
|
7UUN
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with neomycin | | Descriptor: | 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, NEOMYCIN | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Osipiuk, J, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2026-02-25 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance
Nat.Chem.Biol., 20, 2024
|
|
7UUO
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA H135A mutant, complex with tobramycin and coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, COENZYME A, ... | | Authors: | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2026-02-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance
Nat.Chem.Biol., 20, 2024
|
|
8BA7
 
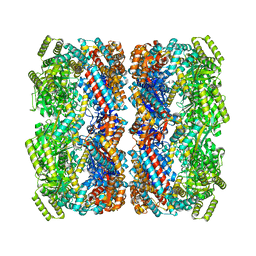 | |
8BA8
 
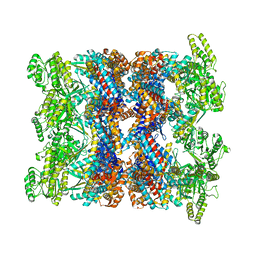 | | CryoEM structure of GroEL-ADP.BeF3-Rubisco. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Gardner, S, Saibil, H.R. | | Deposit date: | 2022-10-11 | | Release date: | 2023-12-27 | | Last modified: | 2026-02-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of substrate progression through the bacterial chaperonin cycle.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8BA9
 
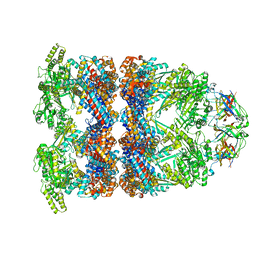 | | CryoEM structure of GroEL-GroES-ADP.AlF3-Rubisco. | | Descriptor: | 60 kDa chaperonin, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Gardner, S, Saibil, H.R. | | Deposit date: | 2022-10-11 | | Release date: | 2023-12-27 | | Last modified: | 2026-02-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of substrate progression through the bacterial chaperonin cycle.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8BAA
 
 | |
8BJ7
 
 | |
8BJ8
 
 | |
8EZR
 
 | | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, native protein | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, HipS(Lp), ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Di Leo, R, Lin, J, Ensminger, A, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-11-01 | | Release date: | 2023-09-27 | | Last modified: | 2026-02-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional diversification despite structural congruence in the HipBST toxin-antitoxin system of Legionella pneumophila.
Mbio, 14, 2023
|
|
