1EAN
 
 | | THE RUNX1 Runt domain at 1.70A resolution: A structural switch and specifically bound chloride ions modulate DNA binding | | Descriptor: | CHLORIDE ION, RUNT-RELATED TRANSCRIPTION FACTOR 1 | | Authors: | Backstrom, S, Wolf-Watz, M, Grundstrom, C, Hard, T, Grundstrom, T, Sauer, U.H. | | Deposit date: | 2001-07-13 | | Release date: | 2002-09-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Runx1 Runt Domain at 1.25 A Resolution: A Structural Switch and Specifically Bound Chloride Ions Modulate DNA Binding
J.Mol.Biol., 322, 2002
|
|
1EAO
 
 | | THE RUNX1 Runt domain at 1.4A resolution: a structural switch and specifically bound chloride ions modulate DNA binding | | Descriptor: | BROMIDE ION, RUNT-RELATED TRANSCRIPTION FACTOR 1 | | Authors: | Backstrom, S, Wolf-Watz, M, Grundstrom, C, Hard, T.H, Grundstrom, T, Sauer, U.H. | | Deposit date: | 2001-07-14 | | Release date: | 2002-09-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Runx1 Runt Domain at 1.25A Resolution: A Structural Switch and Specifically Bound Chloride Ions Modulate DNA Binding
J.Mol.Biol., 322, 2002
|
|
1EAQ
 
 | | The RUNX1 Runt domain at 1.25A resolution: A structural switch and specifically bound chloride ions modulate DNA binding | | Descriptor: | CHLORIDE ION, RUNT-RELATED TRANSCRIPTION FACTOR 1 | | Authors: | Backstrom, S, Wolf-Watz, M, Grundstrom, C, Hard, T, Grundstrom, T, Sauer, U.H. | | Deposit date: | 2001-07-14 | | Release date: | 2002-09-12 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Runx1 Runt Domain at 1.25 A Resolution: A Structural Switch and Specifically Bound Chloride Ions Modulate DNA Binding
J.Mol.Biol., 322, 2002
|
|
1EAR
 
 | | Crystal structure of Bacillus pasteurii UreE at 1.7 A. Type II crystal form. | | Descriptor: | UREASE ACCESSORY PROTEIN UREE, ZINC ION | | Authors: | Remaut, H, Safarov, N, Ciurli, S, Van Beeumen, J. | | Deposit date: | 2001-07-16 | | Release date: | 2002-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Ni2+ Transport and Assembly of the Urease Active Site by the Metallochaperone Uree from Bacillus Pasteurii
J.Biol.Chem., 276, 2001
|
|
1EAS
 
 | | NONPEPTIDIC INHIBITORS OF HUMAN LEUKOCYTE ELASTASE. 3. DESIGN, SYNTHESIS, X-RAY CRYSTALLOGRAPHIC ANALYSIS, AND STRUCTURE-ACTIVITY RELATIONSHIPS FOR A SERIES OF ORALLY ACTIVE 3-AMINO-6-PHENYLPYRIDIN-2-ONE TRIFLUOROMETHYL KETONES | | Descriptor: | 3-[[(METHYLAMINO)SULFONYL]AMINO]-2-OXO-6-PHENYL-N-[3,3,3-TRIFLUORO-1-(1-METHYLETHYL)-2-OXOPHENYL]-1(2H)-PYRIDINE ACETAMIDE, PORCINE PANCREATIC ELASTASE, SODIUM ION, ... | | Authors: | Ceccarelli, C. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nonpeptidic inhibitors of human leukocyte elastase. 3. Design, synthesis, X-ray crystallographic analysis, and structure-activity relationships for a series of orally active 3-amino-6-phenylpyridin-2-one trifluoromethyl ketones.
J.Med.Chem., 37, 1994
|
|
1EAT
 
 | | NONPEPTIDIC INHIBITORS OF HUMAN LEUKOCYTE ELASTASE. 5. DESIGN, SYNTHESIS, AND X-RAY CRYSTALLOGRAPHY OF A SERIES OF ORALLY ACTIVE 5-AMINO-PYRIMIDIN-6-ONE-CONTAINING TRIFLUOROMETHYLKETONES | | Descriptor: | 2-[5-METHANESULFONYLAMINO-2-(4-AMINOPHENYL)-6-OXO-1,6-DIHYDRO-1-PYRIMIDINYL]-N-(3,3,3-TRIFLUORO-1-ISOPROPYL-2-OXOPROPYL)ACETAMIDE, PORCINE PANCREATIC ELASTASE, SODIUM ION, ... | | Authors: | Ceccarelli, C. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nonpeptidic inhibitors of human leukocyte elastase. 5. Design, synthesis, and X-ray crystallography of a series of orally active 5-aminopyrimidin-6-one-containing trifluoromethyl ketones.
J.Med.Chem., 38, 1995
|
|
1EAU
 
 | | NONPEPTIDIC INHIBITORS OF HUMAN LEUKOCYTE ELASTASE. 6. DESIGN OF A POTENT, INTRATRACHEALLY ACTIVE, PYRIDONE-BASED TRIFLUOROMETHYL KETONE | | Descriptor: | 2-[5-AMINO-6-OXO-2-(2-THIENYL)-1,6-DIHYDROPYRIMIDIN-1-YL)-N-[3,3-DIFLUORO -1-ISOPROPYL-2-OXO-3-(N-(2-MORPHOLINO ETHYL)CARBAMOYL]PROPYL]ACETAMIDE, PORCINE PANCREATIC ELASTASE, SODIUM ION, ... | | Authors: | Ceccarelli, C. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nonpeptidic inhibitors of human leukocyte elastase. 6. Design of a potent, intratracheally active, pyridone-based trifluoromethyl ketone.
J.Med.Chem., 38, 1995
|
|
1EAV
 
 | |
1EAY
 
 | | CHEY-BINDING (P2) DOMAIN OF CHEA IN COMPLEX WITH CHEY FROM ESCHERICHIA COLI | | Descriptor: | CHEA, CHEY | | Authors: | Mcevoy, M.M, Hausrath, A.C, Randolph, G.B, Remington, S.J, Dahlquist, F.W. | | Deposit date: | 1998-04-23 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two binding modes reveal flexibility in kinase/response regulator interactions in the bacterial chemotaxis pathway.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1EAZ
 
 | | Crystal structure of the phosphoinositol (3,4)-bisphosphate binding PH domain of TAPP1 from human. | | Descriptor: | CITRIC ACID, TANDEM PH DOMAIN CONTAINING PROTEIN-1 | | Authors: | Thomas, C.C, Dowler, S, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2001-07-17 | | Release date: | 2002-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the Phosphatidylinositol 3,4-Bisphosphate-Binding Pleckstrin Homology (Ph) Domain of Tandem Ph-Domain-Containing Protein 1 (Tapp1): Molecular Basis of Lipid Specificity
Biochem.J., 358, 2001
|
|
1EB0
 
 | | Crystal structure of Bacillus pasteurii UreE at 1.85 A, phased by SIRAS. Type I crystal form. | | Descriptor: | UREASE ACCESSORY PROTEIN UREE, ZINC ION | | Authors: | Remaut, H, Safarov, N, Ciurli, S, Van Beeumen, J. | | Deposit date: | 2001-07-17 | | Release date: | 2002-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Ni2+ Transport and Assembly of the Urease Active Site by the Metallochaperone Uree from Bacillus Pasteurii
J.Biol.Chem., 276, 2001
|
|
1EB3
 
 | | YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE 4,7-DIOXOSEBACIC ACID COMPLEX | | Descriptor: | 4,7-DIOXOSEBACIC ACID, 5-AMINOLAEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Coates, L, Newbold, R, Brindley, A.A, Stauffer, F, Wood, S.P, Warren, M.J, Cooper, J.B, Shoolingin-Jordan, P.M, Neier, R. | | Deposit date: | 2001-07-18 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Two Diacid Inhibitors
FEBS Lett., 503, 2001
|
|
1EB4
 
 | |
1EB6
 
 | | Deuterolysin from Aspergillus oryzae | | Descriptor: | 1,2-ETHANEDIOL, NEUTRAL PROTEASE II, ZINC ION | | Authors: | McAuley, K.E, Jia-Xing, Y, Dodson, E.J, Lehmbeck, J, Ostergaard, P.R, Wilson, K.S. | | Deposit date: | 2001-07-19 | | Release date: | 2001-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A Quick Solution: Ab Initio Structure Determination of a 19 kDa Metalloproteinase Using Acorn
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1EBB
 
 | | Bacillus stearothermophilus YhfR | | Descriptor: | GLYCEROL, PHOSPHATASE, SULFATE ION | | Authors: | Rigden, D.J, Jedrzejas, M.J. | | Deposit date: | 2001-07-24 | | Release date: | 2002-02-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of Action of a Cofactor-Dependent Phosphoglycerate Mutase Homolog from Bacillus Stearothermophilus with Broad Specificity Phosphatase Activity.
J.Mol.Biol., 315, 2002
|
|
1EBC
 
 | | SPERM WHALE MET-MYOGLOBIN:CYANIDE COMPLEX | | Descriptor: | CYANIDE ION, PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Rosano, C, Ascenzi, P, Rizzi, M, Losso, R, Bolognesi, M. | | Deposit date: | 1999-03-04 | | Release date: | 1999-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyanide binding to Lucina pectinata hemoglobin I and to sperm whale myoglobin: an x-ray crystallographic study.
Biophys.J., 77, 1999
|
|
1EBF
 
 | | HOMOSERINE DEHYDROGENASE FROM S. CEREVISIAE COMPLEX WITH NAD+ | | Descriptor: | HOMOSERINE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | DeLaBarre, B, Thompson, P.R, Wright, G.D, Berghuis, A.M. | | Deposit date: | 2000-01-24 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of homoserine dehydrogenase suggest a novel catalytic mechanism for oxidoreductases.
Nat.Struct.Biol., 7, 2000
|
|
1EBM
 
 | | CRYSTAL STRUCTURE OF THE HUMAN 8-OXOGUANINE GLYCOSYLASE (HOGG1) BOUND TO A SUBSTRATE OLIGONUCLEOTIDE | | Descriptor: | 8-OXOGUANINE DNA GLYCOSYLASE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*TP*CP*CP*AP*(8OG)P*GP*TP*CP*TP*AP*CP*C)-3'), ... | | Authors: | Bruner, S.D, Norman, D.P, Verdine, G.L. | | Deposit date: | 2000-01-24 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA.
Nature, 403, 2000
|
|
1EBT
 
 | |
1EBU
 
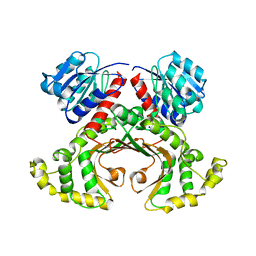 | | HOMOSERINE DEHYDROGENASE COMPLEX WITH NAD ANALOGUE AND L-HOMOSERINE | | Descriptor: | 3-AMINOMETHYL-PYRIDINIUM-ADENINE-DINUCLEOTIDE, HOMOSERINE DEHYDROGENASE, L-HOMOSERINE, ... | | Authors: | DeLaBarre, B, Thompson, P.R, Wright, G.D, Berghuis, A.M. | | Deposit date: | 2000-01-24 | | Release date: | 2000-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of homoserine dehydrogenase suggest a novel catalytic mechanism for oxidoreductases.
Nat.Struct.Biol., 7, 2000
|
|
1EBV
 
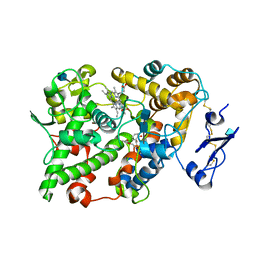 | | OVINE PGHS-1 COMPLEXED WITH SALICYL HYDROXAMIC ACID | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID SALICYLOYL-AMINO-ESTER, ... | | Authors: | Loll, P.J, Sharkey, C.T, O'Connor, S.J, Fitzgerald, D.J. | | Deposit date: | 2000-01-24 | | Release date: | 2000-02-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | O-acetylsalicylhydroxamic acid, a novel acetylating inhibitor of prostaglandin H2 synthase: structural and functional characterization of enzyme-inhibitor interactions.
Mol.Pharmacol., 60, 2001
|
|
1EBW
 
 | | HIV-1 protease in complex with the inhibitor BEA322 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-[DIBENZYL]-GLUCARYL]-DI-[ISOLEUCYL-AMIDO-METHANE] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
1EBY
 
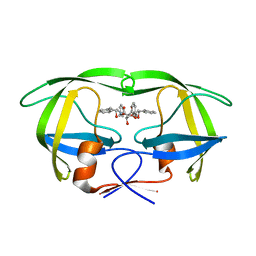 | | HIV-1 protease in complex with the inhibitor BEA369 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-DIBENZYL-GLUCARYL]-DI-[1-AMINO-INDAN-2-OL] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
1EBZ
 
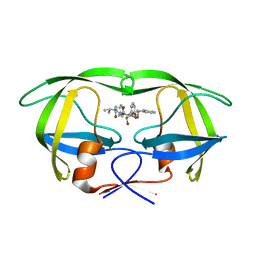 | | HIV-1 protease in complex with the inhibitor BEA388 | | Descriptor: | HIV-1 PROTEASE, [5-(2-HYDROXY-INDAN-1-YLCARBAMOYL)-3,4-DIHYDROXY-2,5-[DIBENZYL-OXY]-PENTANOYL]-VALINYL-AMIDO-METHANE | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
1EC0
 
 | | HIV-1 protease in complex with the inhibitor bea403 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-DI-2-FLUORO-BENZYL-GLUCARYL]-DI-[1-AMINO-INDAN-2-OL] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Symmetric fluoro-substituted diol-based HIV protease inhibitors. Ortho-fluorinated and meta-fluorinated P1/P1'-benzyloxy side groups significantly improve the antiviral activity and preserve binding efficacy
Eur.J.Biochem., 271, 2004
|
|
