1R2B
 
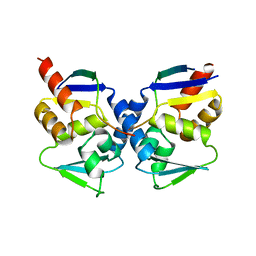 | | Crystal structure of the BCL6 BTB domain complexed with a SMRT co-repressor peptide | | Descriptor: | B-cell lymphoma 6 protein, Nuclear receptor co-repressor 2 | | Authors: | Ahmad, K.F, Melnick, A, Lax, S.A, Bouchard, D, Liu, J, Kiang, C.L, Mayer, S, Licht, J.D, Prive, G.G. | | Deposit date: | 2003-09-26 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain.
Mol.Cell, 12, 2003
|
|
1R2C
 
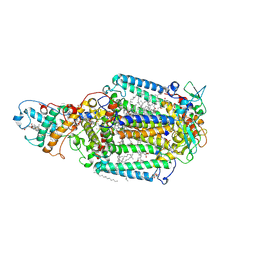 | | PHOTOSYNTHETIC REACTION CENTER BLASTOCHLORIS VIRIDIS (ATCC) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Baxter, R.H, Ponomarenko, N, Pahl, R, Srajer, V, Moffat, K, Norris, J.R. | | Deposit date: | 2003-09-26 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Time-resolved crystallographic studies of light-induced structural changes in the photosynthetic reaction center.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1R2D
 
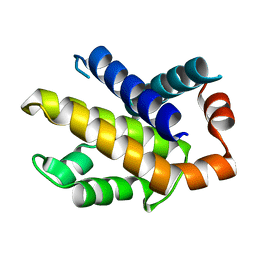 | | Structure of Human Bcl-XL at 1.95 Angstroms | | Descriptor: | Apoptosis regulator Bcl-X | | Authors: | O'Neill, J.W, Manion, M.K, Giedt, C.D, Kim, K.M, Zhang, K.Y, Hockenbery, D.M. | | Deposit date: | 2003-09-26 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Bcl-XL mutations suppress cellular sensitivity to antimycin A.
J.Biol.Chem., 279, 2004
|
|
1R2E
 
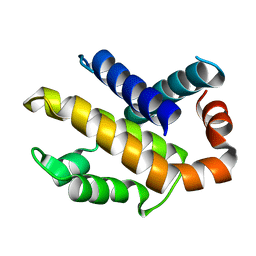 | | Human Bcl-XL containing a Glu to Leu mutation at position 92 | | Descriptor: | Apoptosis regulator Bcl-X | | Authors: | O'Neill, J.W, Manion, M.K, Giedt, C.D, Kim, K.M, Zhang, K.Y, Hockenbery, D.M. | | Deposit date: | 2003-09-26 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bcl-XL mutations suppress cellular sensitivity to antimycin A.
J.Biol.Chem., 279, 2004
|
|
1R2F
 
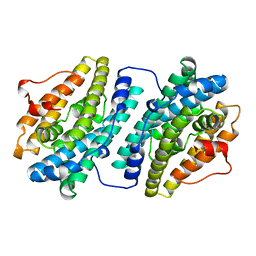 | |
1R2G
 
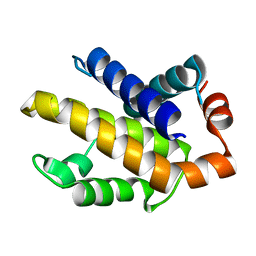 | | Human Bcl-XL containing a Phe to Trp mutation at position 97 | | Descriptor: | Apoptosis regulator Bcl-X | | Authors: | O'Neill, J.W, Manion, M.K, Giedt, C.D, Kim, K.M, Zhang, K.Y, Hockenbery, D.M. | | Deposit date: | 2003-09-26 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Bcl-XL mutations suppress cellular sensitivity to antimycin A.
J.Biol.Chem., 279, 2004
|
|
1R2H
 
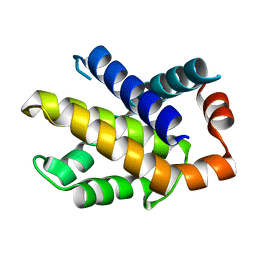 | | Human Bcl-XL containing an Ala to Leu mutation at position 142 | | Descriptor: | Apoptosis regulator Bcl-X | | Authors: | O'Neill, J.W, Manion, M.K, Giedt, C.D, Kim, K.M, Zhang, K.Y, Hockenbery, D.M. | | Deposit date: | 2003-09-26 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bcl-XL mutations suppress cellular sensitivity to antimycin A.
J.Biol.Chem., 279, 2004
|
|
1R2I
 
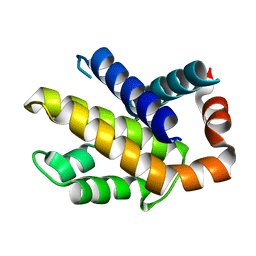 | | Human Bcl-XL containing a Phe to Leu mutation at position 146 | | Descriptor: | Apoptosis regulator Bcl-X | | Authors: | O'Neill, J.W, Manion, M.K, Giedt, C.D, Kim, K.M, Zhang, K.Y, Hockenbery, D.M. | | Deposit date: | 2003-09-26 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bcl-XL mutations suppress cellular sensitivity to antimycin A.
J.Biol.Chem., 279, 2004
|
|
1R2J
 
 | | FkbI for Biosynthesis of Methoxymalonyl Extender Unit of Fk520 Polyketide Immunosuppresant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, protein FkbI | | Authors: | Watanabe, K, Khosla, C, Stroud, R.M, Tsai, S.-C. | | Deposit date: | 2003-09-28 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of an Acyl-ACP Dehydrogenase from the FK520 Polyketide Biosynthetic Pathway: Insights into Extender Unit Biosynthesis
J.Mol.Biol., 334, 2003
|
|
1R2K
 
 | | Crystal structure of MoaB from Escherichia coli | | Descriptor: | Molybdenum cofactor biosynthesis protein B, SULFATE ION | | Authors: | Bader, G, Gomez-Ortiz, M, Haussmann, C, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2003-09-28 | | Release date: | 2004-06-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the molybdenum-cofactor biosynthesis protein MoaB of Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1R2M
 
 | | Atomic resolution structure of the HFBII hydrophobin: a self-assembling amphiphile | | Descriptor: | Hydrophobin II, MANGANESE (II) ION | | Authors: | Hakanpaa, J, Paananen, A, Askolin, S, Nakari-Setala, T, Parkkinen, T, Penttila, M, Linder, M.B, Rouvinen, J. | | Deposit date: | 2003-09-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution structure of the HFBII hydrophobin, a self-assembling amphiphile.
J.Biol.Chem., 279, 2004
|
|
1R2O
 
 | | d(GCATGCT) + Ni2+ | | Descriptor: | 5'-D(*GP*CP*AP*TP*GP*CP*T)-3', NICKEL (II) ION | | Authors: | Cardin, J.C, Gan, Y, Thorpe, J.H, Teixeira, S.C.M, Gale, B.C, Moraes, M.I.A. | | Deposit date: | 2003-09-29 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Metal Ion Distribution and Stabilization of the DNA Quadruplex Structure Formed by d(GCATGCT)
To be published
|
|
1R2Q
 
 | | Crystal Structure of Human Rab5a GTPase Domain at 1.05 A resolution | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Terzyan, S, Zhu, G, Li, G, Zhang, X.C. | | Deposit date: | 2003-09-29 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Refinement of the structure of human Rab5a GTPase domain at 1.05 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1R2R
 
 | | CRYSTAL STRUCTURE OF RABBIT MUSCLE TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Aparicio, R, Ferreira, S.T, Polikarpov, I. | | Deposit date: | 2003-09-29 | | Release date: | 2003-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Closed conformation of the active site loop of rabbit muscle triosephosphate isomerase in the absence of substrate: evidence of conformational heterogeneity.
J.Mol.Biol., 334, 2003
|
|
1R2S
 
 | |
1R2T
 
 | |
1R2Y
 
 | | MutM (Fpg) bound to 8-oxoguanine (oxoG) containing DNA | | Descriptor: | 5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*TP*CP*CP*AP*(8OG)P*GP*TP*CP*TP*AP*CP*C)-3', MutM, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2003-09-30 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | DNA Lesion Recognition by the Bacterial Repair Enzyme MutM.
J.Biol.Chem., 278, 2003
|
|
1R2Z
 
 | | MutM (Fpg) bound to 5,6-dihydrouracil (DHU) containing DNA | | Descriptor: | 5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*TP*CP*CP*AP*(DHU)P*GP*TP*CP*TP*AP*CP*C)-3', MutM, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2003-09-30 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | DNA Lesion Recognition by the Bacterial Repair Enzyme MutM.
J.Biol.Chem., 278, 2003
|
|
1R30
 
 | | The Crystal Structure of Biotin Synthase, an S-Adenosylmethionine-Dependent Radical Enzyme | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, Biotin synthase, ... | | Authors: | Berkovitch, F, Nicolet, Y, Wan, J.T, Jarrett, J.T, Drennan, C.L. | | Deposit date: | 2003-09-30 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme.
Science, 303, 2004
|
|
1R31
 
 | | HMG-CoA reductase from Pseudomonas mevalonii complexed with HMG-CoA | | Descriptor: | (R)-MEVALONATE, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, COENZYME A, ... | | Authors: | Watson, J.M, Steussy, C.N, Burgner, J.W, Lawrence, C.M, Tabernero, L, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 2003-09-30 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Investigations of the Basis for Stereoselectivity from the Binary Complex of HMG-CoA Reductase.
To be Published
|
|
1R33
 
 | | Golgi alpha-mannosidase II complex with 5-thio-D-mannopyranosylamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-thio-alpha-D-mannopyranosylamine, ... | | Authors: | Kuntz, D.A, Xin, W, Kavelekar, L.M, Rose, D.R, Pinto, B.M. | | Deposit date: | 2003-09-30 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 5-Thio-d-glycopyranosylamines and their amidinium salts as potential transition-state mimics of glycosyl hydrolases: synthesis, enzyme inhibitory activities, X-ray crystallography, and molecular modeling
TETRAHEDRON ASYMMETRY, 16, 2005
|
|
1R34
 
 | | Golgi alpha-mannosidase II complex with 5-thio-D-mannopyranosylamidinium salt | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(1Z)-2-phenylethanimidoyl]-5-thio-alpha-D-mannopyranosylamine, ... | | Authors: | Kuntz, D.A, Xin, W, Kavelekar, L.M, Rose, D.R, Pinto, B.M. | | Deposit date: | 2003-09-30 | | Release date: | 2004-11-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 5-Thio-d-glycopyranosylamines and their amidinium salts as potential transition-state mimics of glycosyl hydrolases: synthesis, enzyme inhibitory activities, X-ray crystallography, and molecular modeling
TETRAHEDRON ASYMMETRY, 16, 2005
|
|
1R37
 
 | | Alcohol dehydrogenase from sulfolobus solfataricus complexed with NAD(H) and 2-ethoxyethanol | | Descriptor: | 2-ETHOXYETHANOL, NAD-dependent alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Esposito, L, Bruno, I, Sica, F, Raia, C.A, Giordano, A, Rossi, M, Mazzarella, L, Zagari, A. | | Deposit date: | 2003-09-30 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a ternary complex of the alcohol dehydrogenase from Sulfolobus solfataricus
Biochemistry, 42, 2003
|
|
1R38
 
 | | Crystal structure of H114A mutant of Candida tenuis xylose reductase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, xylose reductase | | Authors: | Kratzer, R, Kavanagh, K.L, Wilson, D.K, Nidetzky, B. | | Deposit date: | 2003-09-30 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Studies of the enzymic mechanism of Candida tenuis xylose reductase (AKR 2B5): X-ray structure and catalytic reaction profile for the H113A mutant
Biochemistry, 43, 2004
|
|
1R39
 
 | | THE STRUCTURE OF P38ALPHA | | Descriptor: | Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Patel, S.B, Cameron, P.M, Frantz-Wattley, B, O'Neill, E, Becker, J.W, Scapin, G. | | Deposit date: | 2003-10-01 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lattice stabilization and enhanced diffraction in human p38 alpha crystals by protein engineering.
Biochim.Biophys.Acta, 1696, 2004
|
|
