1PZP
 
 | |
1PZS
 
 | |
1PZT
 
 | | CRYSTAL STRUCTURE OF W314A-BETA-1,4-GALACTOSYLTRANSFERASE (B4GAL-T1) CATALYTIC DOMAIN WITHOUT SUBSTRATE | | Descriptor: | Beta-1,4-galactosyltransferase 1, SULFATE ION | | Authors: | Ramasamy, V, Ramakrishnan, B, Boeggeman, E, Qasba, P.K. | | Deposit date: | 2003-07-14 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The role of tryptophan 314 in the conformational changes of beta1,4-galactosyltransferase-I
J.Mol.Biol., 331, 2003
|
|
1PZU
 
 | | An asymmetric NFAT1-RHR homodimer on a pseudo-palindromic, Kappa-B site | | Descriptor: | 5'-D(*AP*AP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3', 5'-D(*TP*TP*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*A)-3', Nuclear factor of activated T-cells, ... | | Authors: | Jin, L, Sliz, P, Chen, L, Macian, F, Rao, A, Hogan, P.G, Harrison, S.C. | | Deposit date: | 2003-07-14 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An asymmetric NFAT1 dimer on a pseudo-palindromic KB-like DNA site
Nat.Struct.Biol., 10, 2003
|
|
1PZV
 
 | | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans | | Descriptor: | Probable ubiquitin-conjugating enzyme E2-19 kDa | | Authors: | Schormann, N, Lin, G, Li, S, Symersky, J, Qiu, S, Finley, J, Luo, D, Stanton, A, Carson, M, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-07-14 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans
To be Published
|
|
1PZW
 
 | | Crystal structure of the zinc finger associated domain of the Drosophila transcription factor Grauzone | | Descriptor: | Transcription factor grauzone, ZINC ION | | Authors: | Jauch, R, Bourenkov, G.P, Chung, H.-R, Urlaub, H, Reidt, U, Jaeckle, H, Wahl, M.C. | | Deposit date: | 2003-07-14 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The zinc finger-associated domain of the Drosophila transcription factor grauzone is a novel zinc-coordinating protein-protein interaction module
STRUCTURE, 11, 2003
|
|
1PZX
 
 | | Hypothetical protein APC36103 from Bacillus stearothermophilus: a lipid binding protein | | Descriptor: | Hypothetical protein APC36103, PALMITIC ACID | | Authors: | Zhang, R, Osipiuk, J, Zhou, M, Alkire, R, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-07-14 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lipid binding protein APC36103 from Bacillus Stearothermophilus
To be Published
|
|
1PZY
 
 | | W314A-BETA1,4-GALACTOSYLTRANSFERASE-I COMPLEXED WITH ALPHA-LACTALBUMIN IN THE PRESENCE OF N-ACETYLGLUCOSAMINE, UDP AND MANGANESE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-LACTALBUMIN, BETA-1,4-GALACTOSYLTRANSFERASE, ... | | Authors: | Ramasamy, V, Ramakrishnan, B, Boeggeman, E, Qasba, P.K. | | Deposit date: | 2003-07-14 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Role of Tryptophan 314 in the Conformational Changes of beta 1,4-Galactosyltransferase-I
J.Mol.Biol., 331, 2003
|
|
1PZZ
 
 | | Crystal structure of FGF-1, V51N mutant | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Kim, J, Blaber, M. | | Deposit date: | 2003-07-14 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sequence swapping does not result in conformation swapping for the beta4/beta5 and beta8/beta9 beta-hairpin turns in human acidic fibroblast growth factor
Protein Sci., 14, 2005
|
|
1Q03
 
 | | Crystal structure of FGF-1, S50G/V51G mutant | | Descriptor: | Heparin-binding growth factor 1 | | Authors: | Kim, J, Blaber, M. | | Deposit date: | 2003-07-15 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Sequence swapping does not result in conformation swapping for the beta4/beta5 and beta8/beta9 beta-hairpin turns in human acidic fibroblast growth factor
Protein Sci., 14, 2005
|
|
1Q04
 
 | | Crystal structure of FGF-1, S50E/V51N | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1 | | Authors: | Kim, J, Blaber, M. | | Deposit date: | 2003-07-15 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sequence swapping does not result in conformation swapping for the beta4/beta5 and beta8/beta9 beta-hairpin turns in human acidic fibroblast growth factor
Protein Sci., 14, 2005
|
|
1Q05
 
 | | Crystal structure of the Cu(I) form of E. coli CueR, a copper efflux regulator | | Descriptor: | COPPER (I) ION, Transcriptional regulator cueR | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q06
 
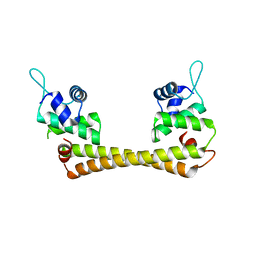 | | Crystal structure of the Ag(I) form of E. coli CueR, a copper efflux regulator | | Descriptor: | SILVER ION, Transcriptional regulator cueR | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q07
 
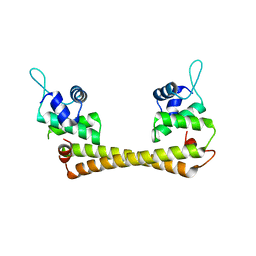 | | Crystal structure of the Au(I) form of E. coli CueR, a copper efflux regulator | | Descriptor: | GOLD ION, Transcriptional regulator cueR | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q08
 
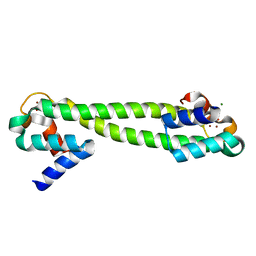 | | Crystal structure of the Zn(II) form of E. coli ZntR, a zinc-sensing transcriptional regulator, at 1.9 A resolution (space group P212121) | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, ZINC ION, ... | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q09
 
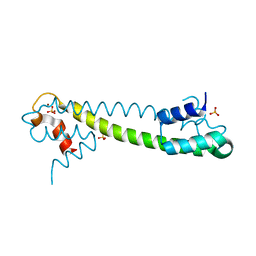 | | Crystal structure of the Zn(II) form of E. coli ZntR, a zinc-sensing transcriptional regulator (space group I4122) | | Descriptor: | SULFATE ION, ZINC ION, Zn(II)-responsive regulator of zntA | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q0A
 
 | | Crystal structure of the Zn(II) form of E. coli ZntR, a zinc-sensing transcriptional regulator (space group C222) | | Descriptor: | SULFATE ION, ZINC ION, Zn(II)-responsive regulator of zntA | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q0C
 
 | | Anerobic Substrate Complex of Homoprotocatechuate 2,3-Dioxygenase from Brevibacterium fuscum. (Complex with 3,4-Dihydroxyphenylacetate) | | Descriptor: | 2-(3,4-DIHYDROXYPHENYL)ACETIC ACID, FE (III) ION, homoprotocatechuate 2,3-dioxygenase | | Authors: | Vetting, M.W, Wackett, L.P, Que, L, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2003-07-15 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases.
J.Bacteriol., 186, 2004
|
|
1Q0D
 
 | | Crystal structure of Ni-containing superoxide dismutase with Ni-ligation corresponding to the oxidized state | | Descriptor: | NICKEL (III) ION, SULFATE ION, Superoxide dismutase [Ni] | | Authors: | Wuerges, J, Lee, J.-W, Yim, Y.-I, Yim, H.-S, Kang, S.-O, Djinovic Carugo, K. | | Deposit date: | 2003-07-16 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of nickel-containing superoxide dismutase reveals another type of active site
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1Q0E
 
 | |
1Q0F
 
 | | Crystal structure of Ni-containing superoxide dismutase with Ni-ligation corresponding to the state after partial x-ray-induced reduction | | Descriptor: | NICKEL (III) ION, SULFATE ION, Superoxide dismutase [Ni] | | Authors: | Wuerges, J, Lee, J.-W, Yim, Y.-I, Yim, H.-S, Kang, S.-O, Djinovic Carugo, K. | | Deposit date: | 2003-07-16 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of nickel-containing superoxide dismutase reveals another type of active site
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1Q0G
 
 | | Crystal structure of Ni-containing superoxide dismutase with Ni-ligation corresponding to the state after full x-ray-induced reduction | | Descriptor: | NICKEL (II) ION, SULFATE ION, Superoxide dismutase [Ni] | | Authors: | Wuerges, J, Lee, J.-W, Yim, Y.-I, Yim, H.-S, Kang, S.-O, Djinovic Carugo, K. | | Deposit date: | 2003-07-16 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of nickel-containing superoxide dismutase reveals another type of active site
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1Q0H
 
 | | Crystal structure of selenomethionine-labelled DXR in complex with fosmidomycin | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, CITRIC ACID, ... | | Authors: | Mac Sweeney, A, Lange, R, D'Arcy, A, Douangamath, A, Surivet, J.-P, Oefner, C. | | Deposit date: | 2003-07-16 | | Release date: | 2004-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of E.coli 1-deoxy-D-xylulose-5-phosphate reductoisomerase in a ternary complex with the antimalarial compound fosmidomycin and NADPH reveals a tight-binding closed enzyme conformation.
J.Mol.Biol., 345, 2005
|
|
1Q0K
 
 | | Crystal structure of Ni-containing superoxide dismutase with Ni-ligation corresponding to the thiosulfate-reduced state | | Descriptor: | NICKEL (II) ION, SULFATE ION, Superoxide dismutase [Ni], ... | | Authors: | Wuerges, J, Lee, J.-W, Yim, Y.-I, Yim, H.-S, Kang, S.-O, Djinovic Carugo, K. | | Deposit date: | 2003-07-16 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of nickel-containing superoxide dismutase reveals another type of active site
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1Q0L
 
 | | Crystal structure of DXR in complex with fosmidomycin | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mac Sweeney, A, Lange, R, D'Arcy, A, Douangamath, A, Surivet, J.-P, Oefner, C. | | Deposit date: | 2003-07-16 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The crystal structure of E.coli 1-deoxy-D-xylulose-5-phosphate reductoisomerase in a ternary complex with the antimalarial compound fosmidomycin and NADPH reveals a tight-binding closed enzyme conformation.
J.Mol.Biol., 345, 2005
|
|
