1P6F
 
 | | Structure of the human natural cytotoxicity receptor NKp46 | | Descriptor: | natural cytotoxicity triggering receptor 1 | | Authors: | Foster, C.E, Colonna, M, Sun, P.D. | | Deposit date: | 2003-04-29 | | Release date: | 2003-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the human natural killer (NK) cell activating receptor NKp46 reveals structural relationship to other leukocyte receptor complex immunoreceptors.
J.Biol.Chem., 278, 2003
|
|
1P6H
 
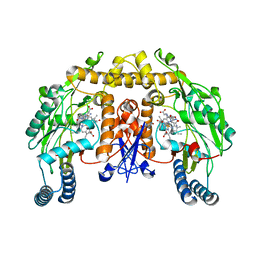 | | Rat neuronal NOS heme domain with L-N(omega)-nitroarginine-2,4-L-diaminobutyric amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, L-N(OMEGA)-NITROARGININE-2,4-L-DIAMINOBUTYRIC AMIDE, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1P6I
 
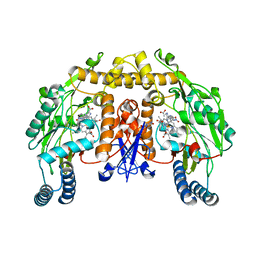 | | Rat neuronal NOS heme domain with (4S)-N-(4-amino-5-[aminoethyl]aminopentyl)-N'-nitroguanidine bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(4S)-4-AMINO-5-[(2-AMINOETHYL)AMINO]PENTYL}-N'-NITROGUANIDINE, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1P6J
 
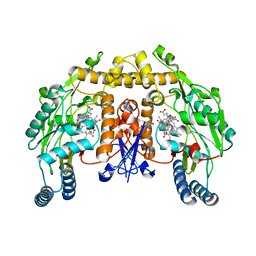 | | Rat neuronal NOS heme domain with L-N(omega)-nitroarginine-(4R)-amino-L-proline amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, L-N(OMEGA)-NITROARGININE-(4R)-AMINO-L-PROLINE AMIDE, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1P6K
 
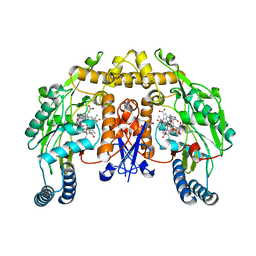 | | Rat neuronal NOS D597N mutant heme domain with L-N(omega)-nitroarginine-2,4-L-diaminobutyric amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1P6L
 
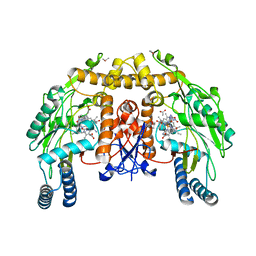 | | Bovine endothelial NOS heme domain with L-N(omega)-nitroarginine-2,4-L-diaminobutyric amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1P6M
 
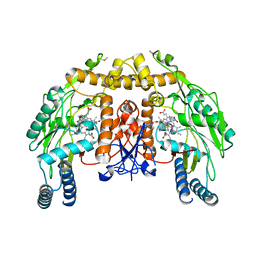 | | Bovine endothelial NOS heme domain with (4S)-N-(4-amino-5-[aminoethyl]aminopentyl)-N'-nitroguanidine bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1P6N
 
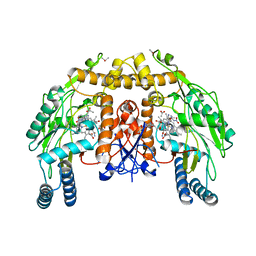 | | Bovine endothelial NOS heme domain with L-N(omega)-nitroarginine-(4R)-amino-L-proline amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1P6O
 
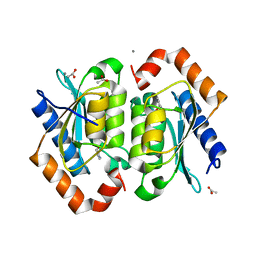 | | The crystal structure of yeast cytosine deaminase bound to 4(R)-hydroxyl-3,4-dihydropyrimidine at 1.14 angstroms. | | Descriptor: | 4-HYDROXY-3,4-DIHYDRO-1H-PYRIMIDIN-2-ONE, ACETIC ACID, CALCIUM ION, ... | | Authors: | Ireton, G.C, Black, M.E, Stoddard, B.L. | | Deposit date: | 2003-04-29 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | The 1.14 a crystal structure of yeast Cytosine deaminase. Evolution of nucleotide salvage enzymes and implications for genetic chemotherapy.
Structure, 11, 2003
|
|
1P6V
 
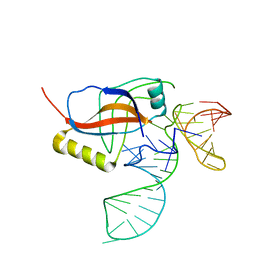 | | Crystal structure of the tRNA domain of transfer-messenger RNA in complex with SmpB | | Descriptor: | 45-MER, SsrA-binding protein | | Authors: | Gutmann, S, Haebel, P.W, Metzinger, L, Sutter, M, Felden, B, Ban, N. | | Deposit date: | 2003-04-30 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB
Nature, 424, 2003
|
|
1P6W
 
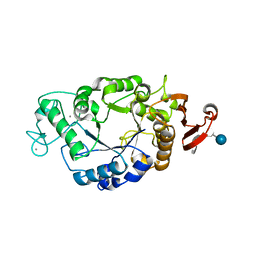 | | Crystal structure of barley alpha-amylase isozyme 1 (AMY1) in complex with the substrate analogue, methyl 4I,4II,4III-tri-thiomaltotetraoside (thio-DP4) | | Descriptor: | CALCIUM ION, PROTEIN (Alpha-amylase type A isozyme), alpha-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose, ... | | Authors: | Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2003-04-30 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of barley alpha-amylase isozyme 1 reveals a novel role of domain C in substrate recognition and binding: a pair of sugar tongs
Structure, 11, 2003
|
|
1P6X
 
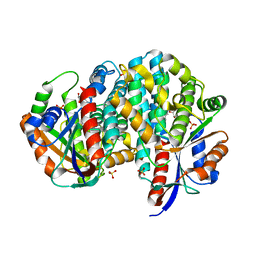 | | Crystal structure of EHV4-TK complexed with Thy and SO4 | | Descriptor: | SULFATE ION, THYMIDINE, Thymidine kinase | | Authors: | Gardberg, A, Shuvalova, L, Monnerjahn, C, Konrad, M, Lavie, A. | | Deposit date: | 2003-04-30 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the dual thymidine and thymidylate kinase activity of herpes thymidine kinases.
Structure, 11, 2003
|
|
1P6Y
 
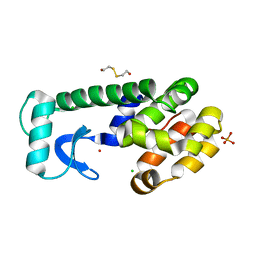 | | T4 LYSOZYME CORE REPACKING MUTANT M120Y/TA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-04-30 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Repacking the Core of T4 Lysozyme by Automated Design
J.Mol.Biol., 332, 2003
|
|
1P71
 
 | | Anabaena HU-DNA corcrystal structure (TR3) | | Descriptor: | 5'-D(*TP*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*CP*AP*CP*C)-3', DNA-binding protein HU | | Authors: | Swinger, K.S, Lemberg, K.M, Zhang, Y, Rice, P.A. | | Deposit date: | 2003-04-30 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Flexible DNA bending in HU-DNA cocrystal structures
Embo J., 22, 2003
|
|
1P72
 
 | | Crystal structure of EHV4-TK complexed with Thy and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, THYMIDINE, ... | | Authors: | Gardberg, A, Shuvalova, L, Monnerjahn, C, Konrad, M, Lavie, A. | | Deposit date: | 2003-04-30 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the dual thymidine and thymidylate kinase activity of herpes thymidine kinases.
Structure, 11, 2003
|
|
1P73
 
 | | Crystal structure of EHV4-TK complexed with TP4A | | Descriptor: | P1-(5'-ADENOSYL)P4-(5'-(2'-DEOXY-THYMIDYL))TETRAPHOSPHATE, SULFATE ION, Thymidine kinase | | Authors: | Gardberg, A, Shuvalova, L, Monnerjahn, C, Konrad, M, Lavie, A. | | Deposit date: | 2003-04-30 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the dual thymidine and thymidylate kinase activity of herpes thymidine kinases.
Structure, 11, 2003
|
|
1P75
 
 | | Crystal structure of EHV4-TK complexed with TP5A | | Descriptor: | P1-(5'-ADENOSYL)P5-(5'-THYMIDYL)PENTAPHOSPHATE, SULFATE ION, Thymidine kinase | | Authors: | Gardberg, A, Shuvalova, L, Monnerjahn, C, Konrad, M, Lavie, A. | | Deposit date: | 2003-04-30 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural basis for the dual thymidine and thymidylate kinase activity of herpes thymidine kinases.
Structure, 11, 2003
|
|
1P78
 
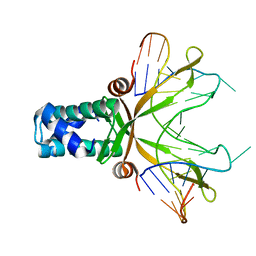 | | Anabaena HU-DNA cocrystal structure (AHU2) | | Descriptor: | 5'-D(*TP*GP*CP*AP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*CP*AP*CP*C)-3', DNA-binding protein HU | | Authors: | Swinger, K.S, Lemberg, K.M, Zhang, Y, Rice, P.A. | | Deposit date: | 2003-04-30 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Flexible DNA bending in HU-DNA cocrystal structures
Embo J., 22, 2003
|
|
1P79
 
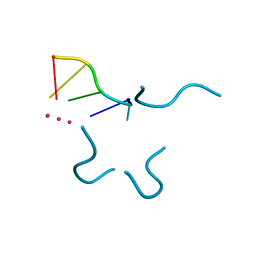 | | Crystal structure of a bulged RNA tetraplex: implications for a novel binding site in RNA tetraplex | | Descriptor: | 5'-R(*UP*GP*UP*GP*GP*U)-3', POTASSIUM ION | | Authors: | Pan, B, Xiong, Y, Shi, K, Sundaralingam, M. | | Deposit date: | 2003-04-30 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of a Bulged RNA Tetraplex at 1.1 A Resolution: Implications for a Novel Binding Site in RNA Tetraplex
STRUCTURE, 11, 2003
|
|
1P7B
 
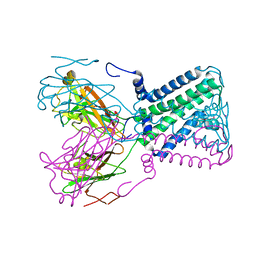 | | Crystal structure of an inward rectifier potassium channel | | Descriptor: | POTASSIUM ION, integral membrane channel and cytosolic domains | | Authors: | Kuo, A, Gulbis, J.M, Antcliff, J.F, Rahman, T, Lowe, E.D, Zimmer, J, Cuthbertson, J, Ashcroft, F.M, Ezaki, T, Doyle, D.A. | | Deposit date: | 2003-05-01 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Crystal structure of the potassium channel KirBac1.1 in the closed state.
Science, 300, 2003
|
|
1P7C
 
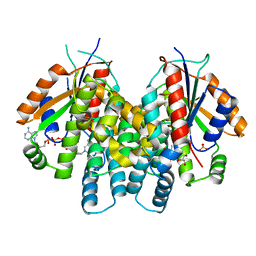 | | Crystal Structure of HSV1-TK complexed with TP5A | | Descriptor: | P1-(5'-ADENOSYL)P5-(5'-THYMIDYL)PENTAPHOSPHATE, SULFATE ION, THYMIDINE, ... | | Authors: | Gardberg, A, Shuvalova, L, Monnerjahn, C, Konrad, M, Lavie, A. | | Deposit date: | 2003-05-01 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the dual thymidine and thymidylate kinase activity of herpes thymidine kinases.
Structure, 11, 2003
|
|
1P7D
 
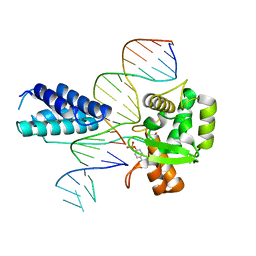 | | Crystal structure of the Lambda Integrase (residues 75-356) bound to DNA | | Descriptor: | 26-MER, 5'-D(*CP*AP*AP*TP*GP*CP*CP*AP*AP*CP*TP*TP*T)-3', Integrase | | Authors: | Aihara, H, Kwon, H.J, Nunes-Duby, S.E, Landy, A, Ellenberger, T. | | Deposit date: | 2003-05-01 | | Release date: | 2003-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A Conformational Switch Controls the DNA Cleavage Activity of Lambda Integrase
Mol.Cell, 12, 2003
|
|
1P7G
 
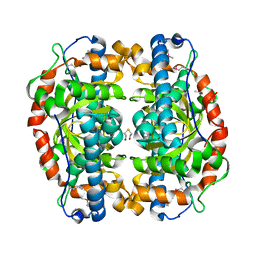 | | Crystal structure of superoxide dismutase from Pyrobaculum aerophilum | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, Superoxide dismutase | | Authors: | Lee, S, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2003-05-01 | | Release date: | 2003-12-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of superoxide dismutase from Pyrobaculum aerophilum presents a challenging case in molecular replacement with multiple molecules, pseudo-symmetry and twinning.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1P7H
 
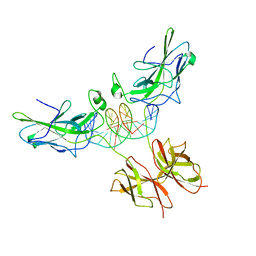 | | Structure of NFAT1 bound as a dimer to the HIV-1 LTR kB element | | Descriptor: | 5'-D(*AP*AP*TP*GP*GP*GP*GP*AP*CP*TP*TP*TP*CP*CP*A)-3', 5'-D(*TP*TP*TP*GP*GP*AP*AP*AP*GP*TP*CP*CP*CP*CP*A)-3', Nuclear factor of activated T-cells, ... | | Authors: | Giffin, M.J, Stroud, J.C, Bates, D.L, von Koenig, K.D, Hardin, J, Chen, L. | | Deposit date: | 2003-05-01 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of NFAT1 bound as a dimer to the HIV-1 LTR kappa B element
Nat.Struct.Biol., 10, 2003
|
|
1P7I
 
 | | CRYSTAL STRUCTURE OF ENGRAILED HOMEODOMAIN MUTANT K52A | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Segmentation polarity homeobox protein engrailed | | Authors: | Stollar, E.J, Mayor, U, Lovell, S.C, Federici, L, Freund, S.M, Fersht, A.R, Luisi, B.F. | | Deposit date: | 2003-05-02 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Engrailed Homeodomain Mutants: IMPLICATIONS FOR STABILITY AND DYNAMICS
J.Biol.Chem., 278, 2003
|
|
