1NN2
 
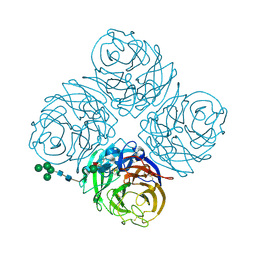 | | THREE-DIMENSIONAL STRUCTURE OF THE NEURAMINIDASE OF INFLUENZA VIRUS A(SLASH)TOKYO(SLASH)3(SLASH)67 AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Varghese, J.N, Colman, P.M. | | Deposit date: | 1991-03-28 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of the neuraminidase of influenza virus A/Tokyo/3/67 at 2.2 A resolution.
J.Mol.Biol., 221, 1991
|
|
1NN3
 
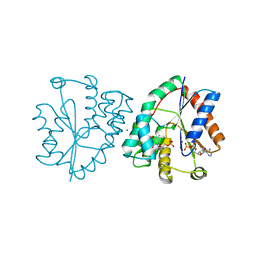 | | Crystal structure of human thymidylate kinase with d4TMP + ADP | | Descriptor: | 3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostermann, N, Segura-Pena, D, Meier, C, Veit, T, Monnerjahn, M, Konrad, M, Lavie, A. | | Deposit date: | 2003-01-12 | | Release date: | 2003-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of human thymidylate kinase in complex with prodrugs:
implications for the structure-based design of novel compounds
Biochemistry, 42, 2003
|
|
1NN5
 
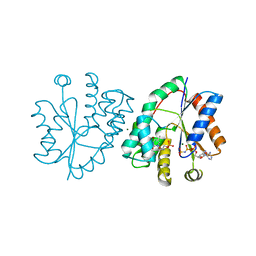 | | Crystal structure of human thymidylate kinase with d4TMP + AppNHp | | Descriptor: | 3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Ostermann, N, Segura-Pena, D, Meier, C, Veit, T, Monnerjahn, M, Konrad, M, Lavie, A. | | Deposit date: | 2003-01-12 | | Release date: | 2003-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of human thymidylate kinase in complex with prodrugs:
implications for the structure-based design of novel compounds
Biochemistry, 42, 2003
|
|
1NN6
 
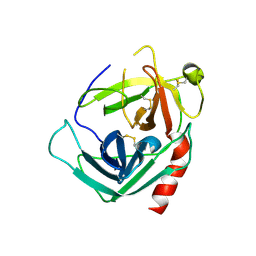 | | Human Pro-Chymase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chymase | | Authors: | Reiling, K.K, Krucinski, J, Miercke, L.J.W, Raymond, W.W, Caughey, G.H, Stroud, R.M. | | Deposit date: | 2003-01-12 | | Release date: | 2003-03-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of human pro-chymase: a model for the activating transition of granule-associated proteases.
Biochemistry, 42, 2003
|
|
1NN7
 
 | |
1NND
 
 | | Arginine 116 is Essential for Nucleic Acid Recognition by the Fingers Domain of Moloney Murine Leukemia Virus Reverse Transcriptase | | Descriptor: | Reverse Transcriptase | | Authors: | Crowther, R.L, Remeta, D.P, Minetti, C.A, Das, D, Montano, S.P, Georgiadis, M.M. | | Deposit date: | 2003-01-13 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and energetic characterization of nucleic acid-binding to the fingers domain of Moloney murine leukemia virus reverse transcriptase
Proteins, 57, 2004
|
|
1NNE
 
 | | Crystal Structure of the MutS-ADPBeF3-DNA complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*GP*CP*GP*AP*CP*GP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*CP*TP*CP*GP*TP*C)-3', 5'-D(P*GP*GP*AP*CP*GP*AP*GP*CP*CP*GP*CP*CP*GP*CP*TP*AP*GP*CP*GP*TP*CP*G)-3', ... | | Authors: | Alani, E, Lee, J.Y, Schofield, M.J, Kijas, A.W, Hsieh, P, Yang, W. | | Deposit date: | 2003-01-13 | | Release date: | 2003-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure and biochemical analysis of the MutS-ADP-Beryllium Fluoride complex
suggests a conserved mechanism for ATP interactions in mismatch repair
J.Biol.Chem., 278, 2003
|
|
1NNF
 
 | | Crystal Structure Analysis of Haemophlius Influenzae Ferric-ion Binding Protein H9Q Mutant Form | | Descriptor: | FE (III) ION, Iron-utilization periplasmic protein, {[-(BIS-CARBOXYMETHYL-AMINO)-ETHYL]-CARBOXYMETHYL-AMINO}-ACETIC ACID | | Authors: | Shouldice, S.R, Dougan, D.R, Skene, R.J, Tari, L.W, McRee, D.E, Yu, R.-H, Schryvers, A.B. | | Deposit date: | 2003-01-13 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High Resolution Structure of an Alternate Form of the Ferric ion Binding Protein from Haemophilus influenzae
J.Biol.Chem., 278, 2003
|
|
1NNH
 
 | | Hypothetical protein from Pyrococcus furiosus Pfu-1801964 | | Descriptor: | SODIUM ION, asparaginyl-tRNA synthetase-related peptide | | Authors: | Tempel, W, Liu, Z.-J, Schubot, F.D, Shah, A, Arendall III, W.B, Rose, J.P, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-01-13 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Hypothetical protein from Pyrococcus furiosus Pfu-1801964
To be published
|
|
1NNI
 
 | | Azobenzene Reductase from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical protein yhda | | Authors: | Cuff, M.E, Kim, Y, Maj, L, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-13 | | Release date: | 2003-07-29 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Azobenzene Reductase from Bacillus subtilis
To be Published, 2003
|
|
1NNJ
 
 | | Crystal structure Complex between the Lactococcus lactis Fpg and an abasic site containing DNA | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*(PDI)P*TP*TP*TP*CP*TP*CP*G)-3', 5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3', Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Serre, L, Pereira de Jesus, K, Boiteux, S, Zelwer, C, Castaing, B. | | Deposit date: | 2003-01-14 | | Release date: | 2003-02-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA.
Nucleic Acids Res., 33, 2005
|
|
1NNK
 
 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-ATPA at 1.85 A resolution. Crystallization with zinc ions. | | Descriptor: | 3-(5-TERT-BUTYL-3-OXIDOISOXAZOL-4-YL)-L-ALANINATE, CHLORIDE ION, Glutamate receptor 2, ... | | Authors: | Lunn, M.-L, Hogner, A, Stensbol, T.B, Gouaux, E, Egebjerg, J, Kastrup, J.S. | | Deposit date: | 2003-01-14 | | Release date: | 2003-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-Dimensional Structure of the Ligand-Binding
Core of GluR2 in Complex with the Agonist (S)-ATPA:
Implications for Receptor Subunit Selectivity.
J.Med.Chem., 46, 2003
|
|
1NNL
 
 | | Crystal structure of Human Phosphoserine Phosphatase | | Descriptor: | CALCIUM ION, CHLORIDE ION, L-3-phosphoserine phosphatase | | Authors: | Peeraer, Y, Rabijns, A, Verboven, C, Collet, J.F, Van Schaftingen, E, De Ranter, C. | | Deposit date: | 2003-01-14 | | Release date: | 2003-06-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | High-resolution structure of human phosphoserine phosphatase in open conformation.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NNO
 
 | | CONFORMATIONAL CHANGES OCCURRING UPON NO BINDING IN NITRITE REDUCTASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | HEME C, HEME D, NITRIC OXIDE, ... | | Authors: | Nurizzo, D, Tegoni, M, Cambillau, C. | | Deposit date: | 1998-07-20 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Conformational changes occurring upon reduction and NO binding in nitrite reductase from Pseudomonas aeruginosa.
Biochemistry, 37, 1998
|
|
1NNQ
 
 | | rubrerythrin from Pyrococcus furiosus Pfu-1210814 | | Descriptor: | Rubrerythrin, ZINC ION | | Authors: | Liu, Z.-J, Tempel, W, Schubot, F.D, Shah, A, Arendall III, W.B, Rose, J.P, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-01-14 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural genomics of Pyrococcus furiosus: X-ray crystallography reveals 3D domain swapping in rubrerythrin
Proteins, 57, 2004
|
|
1NNR
 
 | | Crystal structure of a probable fosfomycin resistance protein (PA1129) from Pseudomonas aeruginosa with sulfate present in the active site | | Descriptor: | MANGANESE (II) ION, SULFATE ION, probable fosfomycin resistance protein | | Authors: | Rife, C.L, Pharris, R.E, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2003-01-14 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Phosphonoformate: a minimal transition state analogue inhibitor of the fosfomycin resistance protein, FosA.
Biochemistry, 43, 2004
|
|
1NNS
 
 | | L-asparaginase of E. coli in C2 space group and 1.95 A resolution | | Descriptor: | ASPARTIC ACID, L-asparaginase II | | Authors: | Sanches, M, Barbosa, J.A.R.G, de Oliveira, R.T, Neto, J.A.A, Polikarpov, I. | | Deposit date: | 2003-01-14 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural comparison of Escherichia coli L-asparaginase in two monoclinic space groups.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NNT
 
 | |
1NNW
 
 | |
1NNX
 
 | | Structure of the hypothetical protein ygiW from E. coli. | | Descriptor: | Protein ygiW, SULFATE ION | | Authors: | Lehmann, C, Galkin, A, Pullalarevu, S, Sarikaya, E, Krajewski, W, Lim, K, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-14 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the hypothetical protein ygiW from E. coli.
To be Published
|
|
1NO1
 
 | | Structure of truncated variant of B.subtilis SPP1 phage G39P helicase loader/inhibitor protein | | Descriptor: | replisome organizer | | Authors: | Bailey, S, Sedelnikova, S.E, Mesa, P, Ayora, S, Waltho, J.P, Ashcroft, A.E, Baron, A.J, Alonso, J.C, Rafferty, J.B. | | Deposit date: | 2003-01-15 | | Release date: | 2003-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of Bacillus subtilis SPP1 phage helicase loader protein G39P
J.Biol.Chem., 278, 2003
|
|
1NO3
 
 | | REFINED STRUCTURE OF SOYBEAN LIPOXYGENASE-3 WITH 4-NITROCATECHOL AT 2.15 ANGSTROM RESOLUTION | | Descriptor: | 4-NITROCATECHOL, FE (III) ION, Lipoxygenase-3 | | Authors: | Skrzypczak-Jankun, E, Borbulevych, O.Y, Jankun, J. | | Deposit date: | 2003-01-15 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Soybean lipoxygenase-3 in complex with 4-nitrocatechol.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1NO4
 
 | | Crystal Structure of the pre-assembly scaffolding protein gp7 from the double-stranded DNA bacteriophage phi29 | | Descriptor: | HEAD MORPHOGENESIS PROTEIN | | Authors: | Morais, M.C, Kanamaru, S, Badasso, M.O, Koti, J.S, Owen, B.A.L, McMurray, C.T, Anderson, D.L, Rossmann, M.G. | | Deposit date: | 2003-01-15 | | Release date: | 2003-07-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacteriophage f29 scaffolding protein gp7 before and after prohead assembly
Nat.Struct.Biol., 10, 2003
|
|
1NO5
 
 | | Structure of HI0073 from Haemophilus influenzae, the nucleotide binding domain of the HI0073/HI0074 two protein nucleotidyl transferase. | | Descriptor: | GLYCEROL, Hypothetical protein HI0073, SODIUM ION, ... | | Authors: | Lehmann, C, Pullalarevu, S, Galkin, A, Krajewski, W, Willis, M.A, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-15 | | Release date: | 2004-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of HI0073 from Haemophilus influenzae, the nucleotide-binding domain of a two-protein nucleotidyl transferase
Proteins, 60, 2005
|
|
1NO7
 
 | | Structure of the Large Protease Resistant Upper Domain of VP5, the Major Capsid Protein of Herpes Simplex Virus-1 | | Descriptor: | Major capsid protein | | Authors: | Bowman, B.R, Baker, M.L, Rixon, F.J, Chiu, W, Quiocho, F.A. | | Deposit date: | 2003-01-15 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the herpesvirus major capsid protein
Embo J., 22, 2003
|
|
