1RKW
 
 | | CRYSTAL STRUCTURE OF THE MULTIDRUG BINDING TRANSCRIPTIONAL REPRESSOR QACR BOUND TO PENTAMADINE | | Descriptor: | 1,5-BIS(4-AMIDINOPHENOXY)PENTANE, SULFATE ION, Transcriptional regulator qacR | | Authors: | Murray, D.S, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 2003-11-23 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structures of QacR-diamidine complexes reveal additional multidrug-binding modes and a novel mechanism of drug charge neutralization.
J.Biol.Chem., 279, 2004
|
|
1RKX
 
 | | Crystal Structure at 1.8 Angstrom of CDP-D-glucose 4,6-dehydratase from Yersinia pseudotuberculosis | | Descriptor: | CDP-glucose-4,6-dehydratase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Vogan, E.M, Bellamacina, C, He, X, Liu, H.W, Ringe, D, Petsko, G.A. | | Deposit date: | 2003-11-23 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure at 1.8 A Resolution of CDP-d-Glucose 4,6-Dehydratase from Yersinia pseudotuberculosis
Biochemistry, 43, 2004
|
|
1RKY
 
 | | PPLO + Xe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Guss, J.M, Duff, A.P. | | Deposit date: | 2003-11-24 | | Release date: | 2004-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Using Xenon as a Probe for Dioxygen-binding Sites in Copper Amine Oxidases
J.Mol.Biol., 344, 2004
|
|
1RL0
 
 | | Crystal structure of a new ribosome-inactivating protein (RIP): dianthin 30 | | Descriptor: | Antiviral protein DAP-30 | | Authors: | Fermani, S, Falini, G, Ripamonti, A, Bolognesi, A, Polito, L, Stirpe, F. | | Deposit date: | 2003-11-24 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.4A structure of dianthin 30 indicates a role of surface potential at the active site of type 1 ribosome inactivating proteins
J.Struct.Biol., 149, 2005
|
|
1RL3
 
 | | Crystal structure of cAMP-free R1a subunit of PKA | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, GLYCEROL, cAMP-dependent protein kinase type I-alpha regulatory chain | | Authors: | Wu, J, Brown, S, Xuong, N.-H, Taylor, S.S. | | Deposit date: | 2003-11-24 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RIalpha subunit of PKA: a cAMP-free structure reveals a hydrophobic capping mechanism for docking cAMP into site B.
Structure, 12, 2004
|
|
1RL4
 
 | | Plasmodium falciparum peptide deformylase complex with inhibitor | | Descriptor: | (2R)-2-{[FORMYL(HYDROXY)AMINO]METHYL}HEXANOIC ACID, 2-{N'-[2-(5-AMINO-1-PHENYLCARBAMOYL-PENTYLCARBAMOYL)-HEXYL]-HYDRAZINOMETHYL}-HEXANOIC ACID(5-AMINO-1-PHENYLCARBAMOYL-PENTYL)-AMIDE, COBALT (II) ION, ... | | Authors: | Robien, M.A, Nguyen, K.T, Kumar, A, Hirsh, I, Turley, S, Pei, D, Hol, W.G.J. | | Deposit date: | 2003-11-24 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | An improved crystal form of Plasmodium falciparum peptide deformylase.
Protein Sci., 13, 2004
|
|
1RL8
 
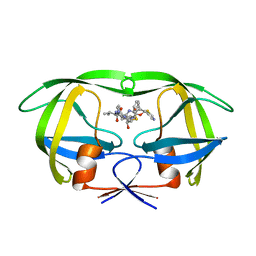 | | Crystal structure of the complex of resistant strain of hiv-1 protease(v82a mutant) with ritonavir | | Descriptor: | RITONAVIR, protease RETROPEPSIN | | Authors: | Rezacova, P, Brynda, J, Sedlacek, J, Konvalinka, J, Fabry, M, Horejsi, M. | | Deposit date: | 2003-11-25 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex of resistant strain of hiv-1 protease(v82a mutant) with ritonavir
To be Published, 2005
|
|
1RL9
 
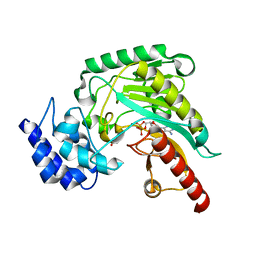 | | Crystal structure of Creatine-ADP arginine kinase ternary complex | | Descriptor: | (DIAMINOMETHYL-METHYL-AMINO)-ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, Arginine kinase, ... | | Authors: | Azzi, A, Clark, S.A, Ellington, R.W, Chapman, M.S. | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The role of phosphagen specificity loops in arginine kinase.
Protein Sci., 13, 2004
|
|
1RLD
 
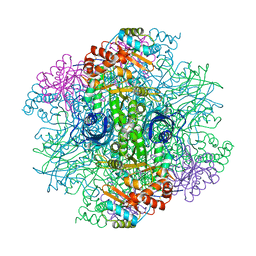 | | SOLID-STATE PHASE TRANSITION IN THE CRYSTAL STRUCTURE OF RIBULOSE 1,5-BIPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE | | Descriptor: | RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN) | | Authors: | Zhang, K.Y.J, Eisenberg, D. | | Deposit date: | 1993-12-10 | | Release date: | 1994-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Solid-state phase transition in the crystal structure of ribulose 1,5-bisphosphate carboxylase/oxygenase.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1RLG
 
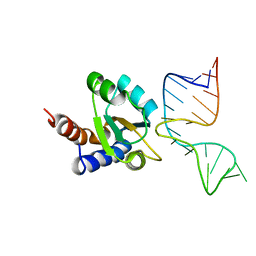 | | Molecular basis of Box C/D RNA-protein interaction: co-crystal structure of the Archaeal sRNP intiation complex | | Descriptor: | 25-MER, 50S ribosomal protein L7Ae | | Authors: | Moore, T, Zhang, Y, Fenley, M.O, Li, H. | | Deposit date: | 2003-11-25 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Basis of Box C/D RNA-Protein Interactions; Cocrystal Structure of Archaeal L7Ae and a Box C/D RNA.
STRUCTURE, 12, 2004
|
|
1RLH
 
 | | Structure of a conserved protein from Thermoplasma acidophilum | | Descriptor: | SODIUM ION, conserved hypothetical protein | | Authors: | Cuff, M.E, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a conserved protein from T. acidophilum
To be Published
|
|
1RLI
 
 | |
1RLJ
 
 | | Structural Genomics, a Flavoprotein NrdI from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, IODIDE ION, NrdI protein | | Authors: | Wu, R, Zhang, R, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-11-25 | | Release date: | 2004-07-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1.5A crystal structure of a thioredoxin-like protein NrdI from Bacillus subtilis
To be Published
|
|
1RLK
 
 | | Structure of Conserved Protein of Unknown Function TA0108 from Thermoplasma acidophilum | | Descriptor: | GLYCEROL, Hypothetical protein Ta0108, SULFATE ION | | Authors: | Osipiuk, J, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-11-25 | | Release date: | 2003-12-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of conserved hypothetical protein TA0108 from Thermoplasma acidophilum
To be Published
|
|
1RLM
 
 | | Crystal Structure of ybiV from Escherichia coli K12 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphatase | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
1RLO
 
 | | Phospho-aspartyl Intermediate Analogue of ybiV from E. coli K12 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphatase | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
1RLT
 
 | | Transition State Analogue of ybiV from E. coli K12 | | Descriptor: | ACETATE ION, ALUMINUM FLUORIDE, GLYCEROL, ... | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
1RLU
 
 | | Mycobacterium tuberculosis FtsZ in complex with GTP-gamma-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Cell division protein ftsZ, GLYCEROL | | Authors: | Leung, A.K.W, White, E.L, Ross, L.J, Reynolds, R.C, DeVito, J.A, Borhani, D.W. | | Deposit date: | 2003-11-26 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches.
J.Mol.Biol., 342, 2004
|
|
1RLV
 
 | |
1RLW
 
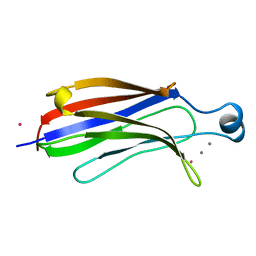 | |
1RLZ
 
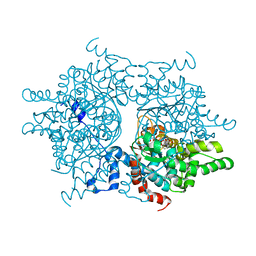 | | Deoxyhypusine synthase holoenzyme in its high ionic strength, low pH crystal form | | Descriptor: | Deoxyhypusine synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Umland, T.C, Wolff, E.C, Park, M.-H, Davies, D.R. | | Deposit date: | 2003-11-26 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A New Crystal Structure of Deoxyhypusine Synthase Reveals the Configuration of the Active Enzyme and of an Enzyme-NAD-Inhibitor Ternary Complex
J.Biol.Chem., 279, 2004
|
|
1RM0
 
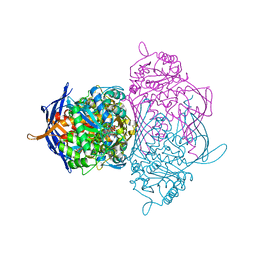 | | Crystal Structure of Myo-Inositol 1-Phosphate Synthase From Saccharomyces cerevisiae In Complex With NAD+ and 2-deoxy-D-glucitol 6-(E)-vinylhomophosphonate | | Descriptor: | (3,4,5,7-TETRAHYDROXY-HEPT-1-ENYL)-PHOSPHONIC ACID, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MANGANESE (II) ION, ... | | Authors: | Jin, X, Foley, K.M, Geiger, J.H. | | Deposit date: | 2003-11-26 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of the 1L-myo-inositol-1-phosphate synthase-NAD+-2-deoxy-D-glucitol 6-(E)-vinylhomophosphonate complex demands a revision of the enzyme mechanism.
J.Biol.Chem., 279, 2004
|
|
1RM1
 
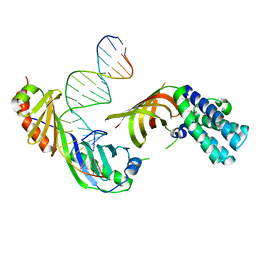 | | Structure of a Yeast TFIIA/TBP/TATA-box DNA Complex | | Descriptor: | 5'-D(*AP*TP*CP*GP*AP*TP*CP*GP*AP*TP*AP*TP*AP*AP*AP*AP*CP*G)-3', 5'-D(P*CP*GP*TP*TP*TP*TP*AP*TP*AP*TP*CP*GP*AP*TP*CP*GP*AP*T)-3', TATA-box binding protein, ... | | Authors: | Jin, X, Gewirth, D.T, Geiger, J.H. | | Deposit date: | 2003-11-26 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High Resolution Structure of a Yeast TFIIA/TBP/TATA-box DNA Complex
TO BE PUBLISHED
|
|
1RM3
 
 | | Crystal structure of mutant T33A of photosynthetic glyceraldehyde-3-phosphate dehydrogenase A4 isoform, complexed with NADP | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase A, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Sparla, F, Fermani, S, Falini, G, Ripamonti, A, Sabatino, P, Pupillo, P, Trost, P. | | Deposit date: | 2003-11-27 | | Release date: | 2004-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Coenzyme Site-directed Mutants of Photosynthetic A(4)-GAPDH Show Selectively Reduced NADPH-dependent Catalysis, Similar to Regulatory AB-GAPDH Inhibited by Oxidized Thioredoxin
J.Mol.Biol., 340, 2004
|
|
1RM4
 
 | | Crystal structure of recombinant photosynthetic glyceraldehyde-3-phosphate dehydrogenase A4 isoform, complexed with NADP | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase A, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Sparla, F, Fermani, S, Falini, G, Ripamonti, A, Sabatino, P, Pupillo, P, Trost, P. | | Deposit date: | 2003-11-27 | | Release date: | 2004-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coenzyme Site-directed Mutants of Photosynthetic A(4)-GAPDH Show Selectively Reduced NADPH-dependent Catalysis, Similar to Regulatory AB-GAPDH Inhibited by Oxidized Thioredoxin
J.Mol.Biol., 340, 2004
|
|
