1OLZ
 
 | | The ligand-binding face of the semaphorins revealed by the high resolution crystal structure of SEMA4D | | Descriptor: | SEMAPHORIN 4D | | Authors: | Love, C.A, Harlos, K, Mavaddat, N, Davis, S.J, Stuart, D.I, Jones, E.Y, Esnouf, R.M. | | Deposit date: | 2003-08-19 | | Release date: | 2003-09-11 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ligand-Binding Face of the Semaphorins Revealed by the High-Resolution Crystal Structure of Sema4D
Nat.Struct.Biol., 10, 2003
|
|
1OM0
 
 | | crystal structure of xylanase inhibitor protein (XIP-I) from wheat | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Xylanase Inhibitor Protein I | | Authors: | Payan, F, Flatman, R, Porciero, S, Williamson, G, Juge, N, Roussel, A. | | Deposit date: | 2003-02-24 | | Release date: | 2003-06-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of xylanase inhibitor protein I (XIP-I), a proteinaceous xylanase inhibitor from wheat (Triticum aestivum, var. Soisson).
Biochem.J., 372, 2003
|
|
1OM4
 
 | | STRUCTURE OF RAT NEURONAL NOS HEME DOMAIN WITH L-ARGININE BOUND | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ARGININE, ... | | Authors: | Li, H, Martasek, P, Shimizu, H, Masters, B.S.S, Poulos, T.L, Raman, C.S. | | Deposit date: | 2003-02-24 | | Release date: | 2003-03-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of Rat Neuronal NOS Heme Domain with L-Arginine Bound
To be Published
|
|
1OM5
 
 | | STRUCTURE OF RAT NEURONAL NOS HEME DOMAIN WITH 3-BROMO-7-NITROINDAZOLE BOUND | | Descriptor: | 3-BROMO-7-NITROINDAZOLE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L, Raman, C.S. | | Deposit date: | 2003-02-24 | | Release date: | 2003-03-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Rat Neuronal NOS Heme Domain with 3-Bromo-7-Nitroindazole Bound
To be Published
|
|
1OM6
 
 | | CRYSTAL STRUCTURE OF A COLD ADAPTED ALKALINE PROTEASE FROM PSEUDOMONAS TAC II 18, CO-CRYSTALLIZED WITH 5mM EDTA (2 MONTHS) | | Descriptor: | CALCIUM ION, SULFATE ION, serralysin | | Authors: | Ravaud, S, Gouet, P, Haser, R, Aghajari, N. | | Deposit date: | 2003-02-25 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the role of divalent metal ions in a bacterial psychrophilic metalloprotease: binding studies of an enzyme in the crystalline state by x-ray crystallography.
J.Bacteriol., 185, 2003
|
|
1OM7
 
 | | CRYSTAL STRUCTURE OF A COLD ADAPTED ALKALINE PROTEASE FROM PSEUDOMONAS TAC II 18, SOAKED IN 85 mM EDTA | | Descriptor: | CALCIUM ION, SERRALYSIN, SULFATE ION | | Authors: | Ravaud, S, Gouet, P, Haser, R, Aghajari, N. | | Deposit date: | 2003-02-25 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Probing the role of divalent metal ions in a bacterial psychrophilic metalloprotease: binding studies of an enzyme in the crystalline state by x-ray crystallography.
J.Bacteriol., 185, 2003
|
|
1OM8
 
 | | CRYSTAL STRUCTURE OF A COLD ADAPTED ALKALINE PROTEASE FROM PSEUDOMONAS TAC II 18, CO-CRYSTALLYZED WITH 10 mM EDTA | | Descriptor: | CALCIUM ION, SERRALYSIN, SULFATE ION | | Authors: | Ravaud, S, Gouet, P, Haser, R, Aghajari, N. | | Deposit date: | 2003-02-25 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the role of divalent metal ions in a bacterial psychrophilic metalloprotease: binding studies of an enzyme in the crystalline state by x-ray crystallography.
J.Bacteriol., 185, 2003
|
|
1OM9
 
 | | Structure of the GGA1-appendage in complex with the p56 binding peptide | | Descriptor: | 15-mer peptide fragment of p56, ADP-ribosylation factor binding protein GGA1 | | Authors: | Collins, B.M, Praefcke, G.J.K, Robinson, M.S, Owen, D.J. | | Deposit date: | 2003-02-25 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for binding of accessory proteins by the appendage domain of GGAs
Nat.Struct.Biol., 10, 2003
|
|
1OMD
 
 | | STRUCTURE OF ONCOMODULIN REFINED AT 1.85 ANGSTROMS RESOLUTION. AN EXAMPLE OF EXTENSIVE MOLECULAR AGGREGATION VIA CA2+ | | Descriptor: | CALCIUM ION, ONCOMODULIN | | Authors: | Ahmed, F.R, Przybylska, M, Rose, D.R, Birnbaum, G.I, Pippy, M.E, Macmanus, J.P. | | Deposit date: | 1990-04-19 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of oncomodulin refined at 1.85 A resolution. An example of extensive molecular aggregation via Ca2+.
J.Mol.Biol., 216, 1990
|
|
1OMH
 
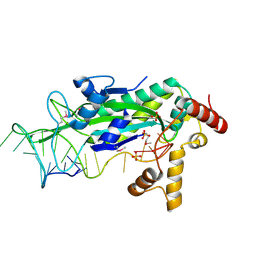 | | Conjugative Relaxase TrwC in complex with OriT Dna. Metal-free structure. | | Descriptor: | DNA OLIGONUCLEOTIDE, SULFATE ION, trwC protein | | Authors: | Guasch, A, Lucas, M, Moncalian, G, Cabezas, M, Perez-Luque, R, Gomis-Ruth, F.X, de la Cruz, F, Coll, M. | | Deposit date: | 2003-02-25 | | Release date: | 2003-11-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC.
Nat.Struct.Biol., 10, 2003
|
|
1OMI
 
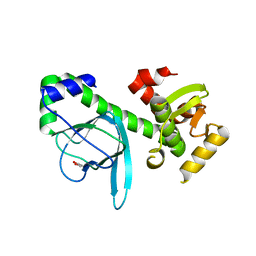 | | Crystal structure of PrfA,the transcriptional regulator in Listeria monocytogenes | | Descriptor: | GLYCEROL, Listeriolysin regulatory protein | | Authors: | Thirumuruhan, R, Rajashankar, K, Fedorov, A.A, Dodatko, T, Chance, M.R, Cossart, P, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-02-25 | | Release date: | 2003-03-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of PrfA, the transcriptional regulator in Listeria monocytogenes
To be Published
|
|
1OMJ
 
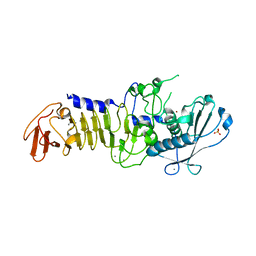 | | CRYSTAL STRUCTURE OF A PSYCHROPHILIC ALKALINE PROTEASE FROM PSEUDOMONAS TAC II 18 | | Descriptor: | CALCIUM ION, SERRALYSIN, SULFATE ION, ... | | Authors: | Ravaud, S, Gouet, P, Haser, R, Aghajari, N. | | Deposit date: | 2003-02-25 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Probing the role of divalent metal ions in a bacterial psychrophilic metalloprotease: binding studies of an enzyme in the crystalline state by x-ray crystallography.
J.Bacteriol., 185, 2003
|
|
1OMK
 
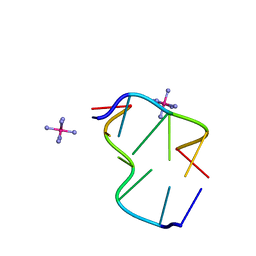 | | The Crystal Structure of d(CACG(5IU)G) | | Descriptor: | 5'-D(*CP*AP*CP*GP*(5IU)P*G)-3', COBALT HEXAMMINE(III) | | Authors: | Schuerman, G, Van Hecke, K, Van Meervelt, L. | | Deposit date: | 2003-02-25 | | Release date: | 2003-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Exploration of the influence of 5-iodo-2'-deoxyuridine incorporation on the structure of d[CACG(IDU)G].
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1OMO
 
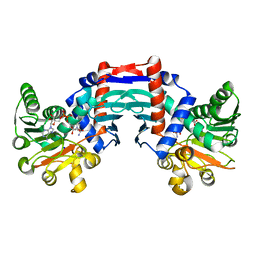 | | alanine dehydrogenase dimer w/bound NAD (archaeal) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, alanine dehydrogenase | | Authors: | Gallagher, D.T, Smith, N.N, Holden, M.J, Schroeder, I, Monbouquette, H.G. | | Deposit date: | 2003-02-25 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of alanine dehydrogenase from Archaeoglobus: active site analysis and relation to bacterial cyclodeaminases and mammalian mu crystallin.
J.Mol.Biol., 342, 2004
|
|
1OMP
 
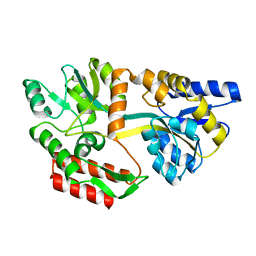 | |
1OMR
 
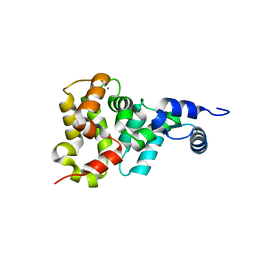 | |
1OMS
 
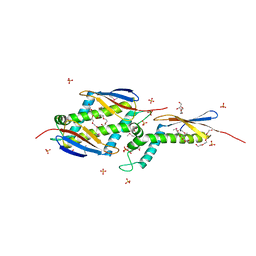 | |
1OMV
 
 | |
1OMW
 
 | | Crystal Structure of the complex between G Protein-Coupled Receptor Kinase 2 and Heterotrimeric G Protein beta 1 and gamma 2 subunits | | Descriptor: | G-protein coupled receptor kinase 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-2 subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(T) beta subunit 1 | | Authors: | Lodowski, D.T, Pitcher, J.A, Capel, W.D, Lefkowitz, R.J, Tesmer, J.J.G. | | Deposit date: | 2003-02-26 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Keeping G proteins at Bay: A Complex Between G Protein-Coupled Receptor Kinase 2 and G-Beta-Gamma
Science, 300, 2003
|
|
1OMX
 
 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2 | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-04-22 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1OMY
 
 | | Crystal Structure of a Recombinant alpha-insect Toxin BmKaIT1 from the scorpion Buthus martensii Karsch | | Descriptor: | ACETIC ACID, Alpha-neurotoxin TX12, CHLORIDE ION | | Authors: | Huang, Y, Huang, Q, Chen, H, Tang, Y, Miyake, H, Kusunoki, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization and preliminary crystallographic study of rBmKalphaIT1, a recombinant alpha-insect toxin from the scorpion Buthus martensii Karsch.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1OMZ
 
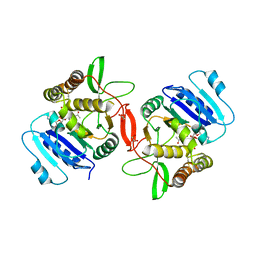 | | crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) in complex with UDPGalNAc | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2),
a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1ON0
 
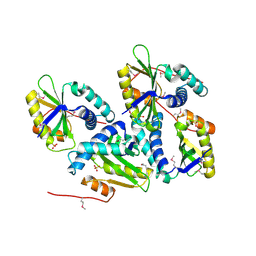 | | Crystal Structure of Putative Acetyltransferase (YycN) from Bacillus subtilis, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR144 | | Descriptor: | CHLORIDE ION, SULFATE ION, YycN protein | | Authors: | Forouhar, F, Shen, J, Kuzin, A, Chiang, Y, Xiao, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-26 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Putative Acetyltransferase (YycN) from Bacillus subtilis
To be Published
|
|
1ON1
 
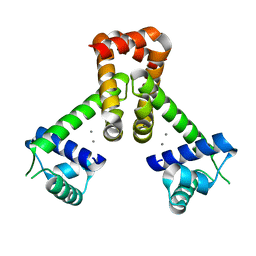 | | Bacillus Subtilis Manganese Transport Regulator (Mntr) Bound To Manganese, AB Conformation. | | Descriptor: | MANGANESE (II) ION, Transcriptional regulator mntR | | Authors: | Glasfeld, A, Guedon, E, Helmann, J.D, Brennan, R.G. | | Deposit date: | 2003-02-26 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Manganese-Bound Manganese Transport Regulator of Bacillus subtilis
Nat.Struct.Biol., 10, 2003
|
|
1ON2
 
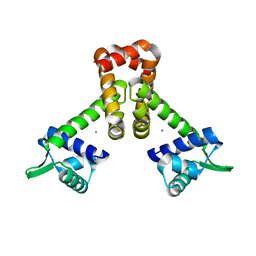 | | Bacillus subtilis Manganese Transport Regulator (MntR), D8M Mutant, Bound to Manganese | | Descriptor: | MANGANESE (II) ION, Transcriptional regulator mntR | | Authors: | Glasfeld, A, Guedon, E, Helmann, J.D, Brennan, R.G. | | Deposit date: | 2003-02-26 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure of the Manganese-Bound Manganese Transport Regulator of Bacillus subtilis
Nat.Struct.Biol., 10, 2003
|
|
